Multiscale Modeling in Computational Biomechanics: Determining Computational Priorities and Addressing Current Challenges
Merryn Tawhai1, Jeff Bischoff2, Daniel Einstein3, Ahmet Erdemir4, Trent Guess5, Jeff Reinbolt6
1Auckland Bioengineering Institute, The University of Auckland, Auckland 1010, NZ
2Zimmer, Inc., PO Box 708, Warsaw, IN 46581-0708, USA
3Biological Monitoring and Modeling, Pacific Northwest National Laboratory, Richland, WA 99352, USA
4Department of Biomedical Engineering, The Cleveland Clinic, Cleveland, OH 44195, USA
5Department of Mechanical Engineering, The University of Missouri, Kansas City, MO 64110, USA
6Department of Bioengineering, Stanford University, Stanford, CA 94305, USA
Corresponding Author: Merryn Tawhai, m.tawhai@auckland.ac.nz
Abstract
In this article, we describe some current multiscale modeling issues in computational biomechanics from the perspective of the musculoskeletal and respiratory systems and mechanotransduction. First, we outline the necessity of multiscale simulations in these biological systems. Then we summarize challenges inherent to multiscale biomechanics modeling, regardless of the subdiscipline, followed by sample computational challenges that are system-specific. We discuss some of the current tools that have been utilized to aid research in multiscale mechanics simulations, and the priorities to further the field of multiscale biomechanics computation.
Keywords (Up to 12 words)
multiscale modeling, respiratory system, musculoskeletal biomechanics, mechanotransduction, finite element analysis, tissue mechanics, cell mechanics
Introduction
Biomechanics is broadly defined as the scientific discipline which investigates the effects of forces acting on and within biological structures. The realm of biomechanics includes the circulatory and respiratory systems, tissue mechanics and mechanotransduction, and the musculoskeletal system and motor control. As in many other biological phenomena, many spatial scales are crossed by biomechanics research: intracellular, multi-cell and extracellular matrix, tissue, organ, and multi-organ systems. It is well established that the effect of forces at higher scales influence behavior at lower scales and that lower scale properties influence higher scale response. However, computational methods that incorporate these interactions in biomechanics are relatively rare. In general, computational models that include representation of multiple spatial or temporal scales are loosely defined as multiscale. The fact that multiscale modeling is not well defined lends the term to a variety of scenarios within the computational physiology community. In biomechanics, multiscale modeling may mean establishing a hierarchical link between spatial and temporal scales while the output of a larger scale system is passed through a finely detailed representation at a lower scale (e.g. body level movement simulations that provide net joint loading for tissue level stress analysis). In reality, multiscale modeling may require more intricate representation of interactions among scales. A concurrent simulation strategy is inevitable to adequately represent nonlinear associations that have been known for decades [1].
Multiscale modeling has existed for many years in basic science and engineering areas such as mathematics, material science, chemistry, and fluid dynamics. Computational and organizational issues common to all these disciplines have been explored, including but not limited to the standardization of methodology, the necessity of reliable data collection procedures, the need for efficient and accurate algorithms, the lack of coupling tools that address multi-physics phenomena, model and data sharing, and public dissemination. Multiscale biomechanics not only shares all of these common problems but it is also hindered by the restricted amount of data collection possibilities for model development and validation, the large variability in anatomical and functional properties, and the readily nonlinear nature of the underlying physics even at single scales.
In this article we describe some current multiscale modeling issues in computational biomechanics from the perspective of the musculoskeletal and respiratory systems and mechanotransduction. First, we justify the requirement of multiscale simulations in these individual systems. Then we summarize challenges inherent to multiscale biomechanics, followed by system-specific computational challenges. We discuss some of the current tools that have been utilized to aid research in multiscale mechanics simulations, and the priorities to further the field of multiscale biomechanics computation. Overall, our goal is to portray our understanding of the highly complicated and time sensitive discipline so called "the multiscale biomechanics modeling".
Need
Musculoskeletal System Perspective
Musculoskeletal modeling can provide the outlining principles of locomotion including movement control and loading on the hard and soft tissues and muscles. Commonly represented at the body level, these models typically use simplified representations of joints (e.g., hip joint as a spherical joint), passive structures (e.g., modeling of ligaments as nonlinear springs), muscles (e.g., hill-type descriptions) and motor control strategies (e.g., calculation of muscle forces using optimization). If the goal is an overall explanation of muscle function and movement at the body level, the added computational and development costs of increasing the level of detail (therefore introducing multiscale modeling) may not be warranted.
There are however cases that warrant multiscale modeling in the analysis of the musculoskeletal system. For example, one may be interested in individual muscle fiber function [2] or the stress-strain profile at the joints [3] during locomotion. There are also scenarios where models of muscle coordination coupled with detailed representation of joints and tissues are needed. In these cases, the interdependency of muscle force and tissue response justifies a concurrent multiscale modeling approach. As an example, patello-femoral pain (PFP) is a common disorder of the knee whose multifactor etiology is not well understood. It is believed that one mechanism of patello-femoral pain is excessive stress in the patellar cartilage. Both muscle activation [4] and muscle reflex response times [5] have been associated with PFP. In addition, the location of pain receptors in the patellar subchondral bone [6], the influence of joint tissues such as the medial patellofemoral ligament [7], and the complexity of calculating cartilage stress, all indicate that a multiscale approach would be beneficial. Temporomandibular joint disorders provide a similar example where complex interdependencies exist between the temporomandibular joint disc and activations of the powerful masticatory muscles [8]. It is generally believed that neuromuscular control is a significant factor in non-contact anterior cruciate ligament (ACL) injury [9]. Understanding non-contact injury mechanisms could be enhanced with multiscale models that include detailed representation of muscle (wrapping, activation, and fiber orientation) coupled with accurate representation of the ACL (interaction with surrounding tissue, insertion areas, fiber orientations, viscoelasticity, and damage accumulation) within a body level framework. Diabetic foot ulceration provides another example where the interaction of muscle coordination and tissue deformation is important. It is well known that diabetic foot ulceration has a mechanical etiology [10]. Patients with diabetes have to perform similar activities of daily living as healthy individuals. The simple task of walking may be harmful since diabetes can affect various levels of biological function from a mechanical perspective. Dysfunctions at these levels manifest themselves in terms of loss of sensation [11], changes in control of movement [12], and alteration of tissue [13] and cell properties [14]. It is not clear how system level mechanical loads, e.g. contact forces at the foot, reflect to cellular deformations that may cause cell damage, therefore ulceration. Higher organ level forces (e.g., increased foot pressures), redistribution of stress due to changes in tissue composition (e.g., muscular atrophy [15]) as well as cell distribution within a tissue, increased mechanical loading of cells or their decreased damage resistance may all have a role in ulceration. A multiscale modeling approach is likely to identify the pathways to cell damage from organ level mechanical loading to cell level deformations.
Holistic simulation of all aforementioned conditions requires models that optimize neuromuscular response concurrently with detailed models of dynamic tissue behavior emphasizing a multiscale approach in musculoskeletal biomechanics. A further requirement is that these multiscale models have sufficient computational efficiency for optimization type simulations.
Respiratory System Perspective
Gas exchange at the respiratory surfaces of the lung is dependent on adequate matching of ventilation and perfusion through complex branching structures that are physically tethered to the surrounding parenchymal tissue. Ventilation, perfusion, and gas exchange are therefore intimately dependent on the relationship between stress and strain in the lung parenchymal, airway, and vascular tissues, how this varies regionally, and how it changes with disease. Fredberg and Kamm [16] recently provided a comprehensive review of stress transmission in the lung, from cell to organ. The review highlights the current state of knowledge of the lung as a mechanical organ with organ-specific interdependencies that arguably make it the most complex system of the human body in which to compute solid mechanics.
The lung parenchymal tissue is extremely delicate, yet is required to undergo relatively large strain during the repeated action of ventilation. The tissue accommodates change in tone of the airways or vasculature through a far lower resistance to shear (and therefore to shape change) than to volume change [17]. The lung tissue deforms readily - due to gravity - with a change of posture, and the resulting regional differences in volume expansion of the lung partly determine the distribution of inspired air. The bronchi and blood vessels are elastic structures that are subjected to internal air and blood pressures, respectively, and through parenchymal tethering they are also subjected to dynamic expanding forces transmitted from the pleural surface. Transmission of force to the airways or vessels depends on the integrity of this tethering: respiratory diseases such as asthma or emphysema disrupt mechanical tethering, but through different mechanisms. Tethering of the arteries and veins extends to the level of the pulmonary capillaries that essentially form the walls of the alveoli. Lung tissue expansion causes expansion of the arteries and veins, but the opposite is true for the capillaries, where tissue expansion at the microstructural level equates to expansion of the alveolar septae (and also potentially some rearrangement of alveolar wall conformation) and results in a reduction in the height of the capillaries. The cyclic motion of the breathing lung also imposes dynamic forces at the cellular level, affecting the regulation of structure, function, and metabolism in a variety of cell types. For example, shear stress via airflow is believed to be transmitted to the epithelial cells, with a role in regulating airway surface liquid, and stretch sensitive ion channels in a number of cell types regulate an increased influx of calcium under mechanical stretch [18]. Research into cellular mechanotransduction - as described in detail in the following section - is providing a wealth of information on the cellular response to stretch or shear, but it is difficult to relate this knowledge to the function of the whole organ without a multi-scale computational framework in which to interpret cell level measurements. The previous examples have considered the influence of the organ/tissue on lower level structures and cell. An example in the 'opposite direction' is the effect of bronchoconstriction on parenchymal mechanics, via tethering of the airway wall to the tissue.
The requirement for multiscale representation in lung mechanics is therefore apparent: from lung interaction with heart, chest wall, and diaphragm, to the organ and its internal structures; from the mechanical behavior of a complex functional tissue and the major role that surface forces play in determining this behavior, down to the level of the variety of lung cells that respond to dynamic mechanical forces.
Mechanotransduction Perspective
The adaptation of tissue properties due to cell function, and mediated by the mechanical and biochemical environment, has long been recognized. The phenomenon of mechanotransduction is associated with many normal and pathologic processes including bone remodeling, cardiovascular development, and wound healing. However, though the roots of the theory of mechanotransduction are over a hundred years old [19], mathematical formulations for the theory that can address the complexities of soft and hard tissue biomechanics have entered into the modeling domain fairly recently.
Continuum manifestation of mechanotransduction is drive by cellular and subcellular events, which immediately suggests two unique modeling strategies: capture the continuum level behavioral characteristics using mathematical laws of growth and modeling, or model fundamental cell-cell interactions on a local level and allow the continuum properties to evolve accordingly. The latter approach has been used in several applications, including mesenchymal morphogenesis [20] and trabecular bone adaptation [21][22]. These analyses are driven by local, relatively simple differential equations that govern the evolution of, for example, bone or cell density, from which continuum level patterns may emerge. The former approach relies on solving continuum-level equilibrium equations, and is reviewed further below. The multiscale nature of this problem, then, is the efficient interfacing between these two approaches.
The effects of mechanotransduction include growth and remodeling, which are typically considered as unique processes [23] - representing mass/volume changes due to bulk material deposition or resorption versus structural changes including trabecular or fiber realignment, respectively. Development of models of these processes involves both constitutive formulation (constructing mathematical models that govern the evolution of the state of the material), and computational implementation of the constitutive model, typically within a finite element framework with a few exceptions [21].
Basic constitutive formulations have been posed in terms of evolution laws for the geometry of a basic construct, for example a cylindrical vessel [24]. In addition to being a continuum formulation for what is inherently a multiscale, multiphysics process, these formulations assume homogeneity of material response. More rigorous constitutive formulations look at the point-wise response of the material, although still in the continuum sense. The continuum mathematical concept of the deformation gradient is decomposed into an elastic component and a growth component [25]. The growth component effectively alters the reference state of the material, and is thus able to capture phenomena such as residual stress/opening angle in vasculature [26] and fiber recruitment/alignment in engineered tendon constructs [27]. The constitutive problem is then to pose the evolution law for the growth component of the deformation gradient in terms of some metric of the local mechanical state including stress [24], strain [28], stiffness [29], or strain energy density [30]. This approach is similar mathematically to nonlinear inelastic constitutive models [31], in which the deformation gradient is decomposed into an elastic component and a viscous/plastic component.
Much of the computational work of geometric/structural adaptation of bone in response to mechanical loading has involved alteration of the mechanical properties (e.g. density, elastic modulus) of a constituent element/voxel in the computational model, and net addition/subtraction of that element/voxel [30]. Computational models of growth/remodeling of soft tissue are also typically finite element based, and can be realized by incorporating user-defined material properties reflective of the inelastic processes into general purpose finite element analysis software [30]. These models are again typically continuum based, employing laws for local element adaptation based on the continuum state.
From a different perspective, mathematical/computational models for cell/matrix interactions have been used to predict cell migration within anisotropic tissue, tissue deformation due to the contractile properties of adherent cells, and fiber reorientation [32]. This approach has been used for hard tissue (bone) to predict remodeling [33] and callus formation [18], and can also be used on conjunction with fiber-based constitutive models for soft tissue behavior in order to predict evolution of material properties due to cell/matrix interactions. However, such an approach is again continuum based.
The case for multi-scale modeling of mechanotransduction is based in the physiological underpinnings of the process itself. Though not yet fully understood, mechanical stimuli are transduced into the cell through structural (integrins that mechanically link the extracellular matrix to the cytoskeleton and in turn the nucleus) and biochemical (stress-based activation of transmembrane ion channels or surface growth factor receptors) pathways [34]. Once internalized, a cascade of intracellular processes ensues, which drives cell function including motility, contraction, proliferation, differentiation, and fibrillogenesis. Continuum-based formulations for mechanotransduction treat these cellular and subcellular processes as 'black boxes'; though such treatment has been shown to successfully capture tissue-level aspects of growth and remodeling, it has been argued that governing the response of cells using mathematical concepts like stress or strain is inherently unsatisfactory [35]. Additionally, mathematical coupling of fundamental cellular processes with their effects on the geometric, structural, and constitutive environment, and in turn continuum-level boundary conditions on cell function can be used to drive functional tissue engineering, understand and better treat pathologic growth/remodeling (hypertrophy), and develop combined mechanical and biological solutions to trauma and joint reconstruction.
Challenges
Using computer models to simulate biological phenomena, regardless of spatial and temporal scales, comes with associated costs in labor and computation. First, any modeling and simulation platform relies on well-developed tools that facilitate defining models and allow simulations to be conducted in reasonable time frames. Developing robust tools is labor intensive and requires expertise in computational science and mathematics. Given model development and numerical solution tools, another labor intensive step relates to development of a model which is directly related to the research discipline. Compiling adequate model input parameters and representing anatomical structures (e.g., generating meshes of complex geometry) are all part of this process. This is usually a hidden cost, not necessarily reported with the results of the studies. Following model development, the next step is to use the model to simulate conditions to answer clinical or research problems. Simulations, particularly in multiscale modeling where coupling between physical domains and scales is necessary, are computationally intensive. Solutions may be obtained but interpretation of them may also be challenging, relying on an expert, possibly spending hours to confirm validity of results and then to extract useful information applicable to the research area. All these general challenges can be addressed in a research setting, if the development cost of tools and models against decreased research output can be afforded. However, the urgent nature of clinical problems increases the burden on model developers, highly demanding easy to use and robust models combined with timely solutions.
Modeling in tissue mechanics requires reliable software to prepare models, to solve them and later to process the results. Preparation of models usually requires data to reconstruct anatomical geometry, to represent characteristics specific to a physical domain of interest, to describe simulation conditions, and for validation. Hardware for model preparation and visualization of results usually requires graphical processors capable of handling large data sets. In general, the simulation process needs hardware with high speed numerical computation capabilities and large memory. It is not uncommon to use high performance computers with multiple processors; parallel processing using shared or distributed memory architectures.
For multiscale modeling and simulations, software, hardware and data needs increase. In general, specific and specialized computational tools are used at each dimensional scale of the musculoskeletal system. Linking and concurrently passing information across scales, each utilizing diverse computational methods, presents a challenge. The primary computational tools of musculoskeletal modeling include multi-body dynamics at the body level, continuum finite element methods at the organ level, and specialized algorithms and solvers at the tissue/cell level. The multi-body method is computationally efficient, but lacks the complexity to accurately capture tissue behavior. Finite element models of organs can estimate tissue deformation, but are typically intensive in both model preparation and computational time. Combinations of measured electromyographic activity (EMG), ground reaction forces, kinematics, musculotendon dynamics models, muscle activation optimization schemes, and forward or inverse dynamics can predict net joint loading and the forces of individual muscles. Joint loading and muscle forces can then provide input to finite element models that calculate tissue deformation. For example, predicted quasi-static muscle forces and relative bone displacements have been used to provide boundary conditions for finite element prediction of cartilage stress [36]. In this scheme, parameters at the organ and tissue level are not part of the muscle control strategy. The finite element method and optimization based force prediction can be coupled [37], but the computational cost of repeatedly solving the finite element model is prohibitive. In addition, unless time history is also passed between the separate computational domains, viscoelastic behavior and contact friction cannot be represented.
At the body level, predictive simulation of musculoskeletal movements is possible by using forward dynamics and optimal control of muscle activations [38]. Such simulations are able to predict muscle control patterns for performance related activities such as maximum height jumping [39], and for efficient movements like walking with minimum energy expenditure [40]. These simulations are already costly due to repeated integration of the equations of motion to solve for an optimal muscular control pattern. Regardless, adding another level of complexity by introducing models at tissue, or even cell scales, have practical implications. For example, one can design rehabilitation strategies that fine tune system level loading to promote healing through cell level remodeling. In addition, safe movement strategies can be predicted to prevent tissue level failures. Concurrent simulations, coupling forward dynamics of a musculoskeletal multi-body system with tissue level finite element models, is possible (Fig. 1). However, this increases the computational cost of movement prediction using single forward dynamics solution from seconds to hours [41]. Remember that the optimal control procedure, which may be seeking minimum tissue stress, can require hundreds if not thousands of these simulations.
Validation of multiscale musculoskeletal models depicts another significant challenge. The current state of in vivo data collection for the musculoskeletal system is comprised of muscle activations, body segment motion, and ground reaction forces. In addition, musculoskeletal organ geometries can be obtained through magnetic resonance imaging (MRI), although high resolution images require in vitro MRI or computed tomography (CT) scans. With the exception of a few limited instances, muscle forces cannot be measured in vivo. Models that involve estimation of muscle force typically verify model predictions through measurement of muscle activation (EMG) [42]. Organ, tissue, and sub-tissue data collection is typically in vitro. This may include mechanical properties of cortical bone, orientation of collagen fibers, and protein content. The lack of in vivo subject specific data and the complexity and cost associated with experimental measurements emphasize the need for data repositories for musculoskeletal biomechanics. Parameter sensitivity studies coupled with statistical populations of in vivo and primarily in vitro data may provide feasible validation routes.
In the study of the mechanics of the lung tissue, the main challenge remains as it stood nearly 25 years ago: “First, stress-strain relations, based on independent material testing or microstructural modeling with some conformational testing, are needed.” [43]. Early computational studies of lung tissue mechanics used linear elastic theory and linear material constants, treating the lung as a uniformly inflated structure subjected to an incremental deformation [44]. The approach was extended by solving for successive small displacement increments, and using elastic moduli that were dependent on the associated incremental change in transmural pressure [45]. In reality the lung undergoes relatively large strain during normal breathing, requiring many of these small increments and potentially accumulating numerical error with each increment, yet the appeal of the linear elastic approach is clear: the governing equations are simpler than equations valid for large deformations, enabling analytic solutions for simple shapes and loading [46], and the elastic moduli are obtained relatively easily [17].
However, an accurate analysis requires the use of governing equations that are valid for large deformations, i.e. finite deformation elasticity. Relatively few studies have used large deformation theory, and all of these have had to compromise to some degree in their constitutive law [47]. The elasticity of the lung parenchyma comprises a tissue contribution and surface forces due to the alveolar air-liquid interface. The magnitude of the surface forces changes with surface area, and surface tension distorts the alveolar geometry as the tissue expands or recoils [48]. A mathematical model of the tissue microstructure that includes interaction between alveolar structure and surface tension has been developed [49], but to date this has not been extended to link with the tissue continuum level. Such a link could be made using the approach presented by [50], where moduli were derived based on an analysis of the elastic response of a model of the lung microstructure. Experimental testing of lung tissue to determine the relationship between stress and strain is technically difficult, and is performed on tissue without its normal surface forces. When there is no surface tension, the tissue component of the parenchymal elasticity is different to that in the intact, in vivo lung because surface tension results in distortion of the tissue causing more elastic energy to be stored. Gathering sufficient accurate data on which to base the derivation of a constitutive law is therefore exceedingly difficult. Ideally an analysis of lung tissue mechanics would also account for viscoelasticity and tissue hysteresis, and the different contributions of – and interaction between - tissue elasticity and surface forces.
Current Tools
Software to develop musculoskeletal models and to simulate/analyze movements is maturing and becoming freely available [51]. These tools promise an open architecture, potentially allowing linking with physiologically realistic simulations of tissue/cell deformations and multiscale muscle models. Finite element analysis packages that focus on biological problems are also provided for free [52], with developers open to implement customization specific to research fields. Nevertheless, current multiscale analysis of the musculoskeletal system is still based on individualization of such tools. Aforementioned examples from other disciplines also illustrate the challenges associated with model preparation and solution. While both of these aspects of multiscale modeling can be computationally intensive, model preparation can, in addition, be quite labor intensive - often to the point where the analyst invests more time in model creation than in model analysis. Overall, this is to the detriment of science. Consider the challenges presented by the linking of scales between the level of the organ and the cell. Organs themselves typically span multiple geometric scales – compare the size of cardiac valves to the coronary arteries, or the size of the trachea to the terminal bronchioles. Efficient grid generation across these geometric scales needs to be somewhat structured so that the physics of both scales are correctly resolved, e.g., the transition from convection-dominated to diffusion-dominated mechanics in the lung. With complex biological domains, this can be daunting. Furthermore, because organs are by definition spatially heterogeneous arrangements of cells, mechanical properties with efficient continuum representations such as tissue elasticity or mass-transfer emerge in a spatially heterogeneous fashion from the cellular and extra-cellular constituents of tissue. Some examples are the spatial arrangement of myofibers in the heart, of collagen and elastin in heart valves and arteries, or of the smooth muscle in the lung. Structural constitutive models, as opposed to phenomenological models, of these continuum fields are highly desirable as they can substantially contract the parameter space. However, this requires the efficient identification and mapping of these cell-level components to a computational grid. In cases where cell activity is modeled together with organ level fields, this mapping must be a two way communication. This latter scenario demands efficient geometric databases that can organize time varying organ and cell level inputs and outputs. Below we outline some current and evolving efforts to meet these challenges.
Several excellent isotropic unstructured tetrahedral grid generation algorithms exist for creating finite-element or finite-volume grids from imaging-derived closed triangulated manifolds, e.g., [53]. Isotropic algorithms attempt to construct tetrahedral elements with nearly equal internal angles and approximately equal edge lengths. However, with medical imaging data, the lengths of these edges are typically more related to the resolution of the image than they are to the physics to be solved and do not take the geometric scale into account. Moreover, the tessellation of the volume is typically disordered. Structured grids, typically structured hexahedral grids, are computationally efficient and can be made to fit the physics of the problem, but structured grids can be laborious to construct if they can be constructed at all. Forecasts for petascale computing envision that a fourfold increase in speedup will come from adaptive gridding, while only a one-and-a-half-fold speedup will come from increased parallelism [54]. These forecasts forcefully apply to large-scale biomedical problems. A compromise between fully disordered tetrahedral grids and structured hexahedral grids are a new class of algorithms that automatically generate structured, scale-invariant tetrahedral grids from medical imaging data [55]. These grids are well suited to large-scale computations as they can be designed to concentrate computational grid where it is needed while keeping the overall computational cost of the problem tractable. They are also structured in the sense that elements can be automatically arranged in a user specified number of nearly orthogonal layers.
The approach (Fig. 2) consists of defining a feature-size field on single or multiple material input triangulated surface mesh derived from single or multiple material Marching Cubes algorithms. This is accomplished without a background grid and without referencing the medial axis. Thus, determination of the feature-size field is not only computationally efficient, but also robust in the sense that it is continuous and does not change unreasonably under perturbation of the surface mesh. Prior to volume mesh generation, the input surface mesh is modified (refined and de-refined) so that edge lengths are proportional to the feature size field, with the constraint that refinement/de-refinement preserves topology and curvature. Surface modification is iterated with a volume-conserving smoothing [56], with the result that surface triangles are well-shaped, well-organized and graded. Volume conserving smoothing also maintains the enclosed volume to within machine precision with respect to the original voxelated volume. From these - possibly multiple - modified surfaces, a user-defined number of structured layers are constructed across arbitrarily oriented cross-sections in the domain, independent of scale. User intervention consists solely of specifying the desired number of layers and the desired element anisotropy. Additionally, a scale-dependent function may be specified. A constant function, for example, would result in a constant number of layers independent of scale. A linear function would proportionally increase or decrease the number of layers at either top or bottom scales. Functions may be completely arbitrary.
Once an efficient computable grid has been defined, it is necessary to establish a bridge between measurements of cell-level data and the organ level model. These data can become the inputs to upscaling approaches in which effective reaction-diffusion equations or elasticity tensors are locally defined by homogenizing cells and extracellular proteins over spatial windows of several cells. Alternatively, they can be stored in the computational cells of the organ-level in order to drive complimentary cell-level models. In either case, some averaging of data inevitably occurs. In the case where these data derive from imaging modalities such as MRI, PET or CT, volume-to-volume mapping can be done efficiently [57], and can be made to tolerate modest geometric mismatching. However, often in multi-scale models, upscaling is used to define boundary conditions. This requires a conservative volume-to-surface mapping. Along these lines, more work needs to be done.
If voxel to unstructured grid mappings are presently feasible, a much greater potential source of cell-level data is histology [58]. However, histology-based data (fluorescence, autoradiography, in-situ hybridization, proteomics, immunohistology, or simple H&E staining) pose two fundamental challenges: reconstruction and quantitation. Reconstruction is challenging because sections that are thin enough to reveal cell-level detail under analysis are also prone to distortion during the process of sectioning. Some recent algorithm development has been dedicated to accurate reconstructions without a reference geometry especially in neuroscience, e.g., [59]. These and related algorithms attempt to affect local nonlinear warp transformations on a slice-by-slice basis in an effort to rectify these distortions. These efforts are promising, but more work is needed to extend and adapt these approaches to other organs such as the heart and lung. Quantitation can be challenging depending on the modality. For example, with in-situ hybridization mRNA for a specific marker is hybridized to the tissue and the signal strength is proportional to the local concentration. The ability to quantitate that signal in at least a semi-quantitative fashion is essentially an image processing problem [60]. Here again, work in neuroscience indicates a possible path for multiscale tissue modeling, where markers can be specialized for, collagen, elastin and muscle fibers, and histology can provide both spatial information and local density [58].
Once these data have been referenced to a single geometry, whether voxelated or not, they may be efficiently mapped to unstructured grids with the same methods as are applied to MR or CT data [57]. However, the sheer volume of data represented by histological data with near cellular resolution is prohibitive for efficient communication between the computational grid and the three-dimensional database. A preferable solution is to adopt a multi-resolution, grid-based approach [61], allowing the resolution of the 3D database to be adapted to the averaging window that works best for the communication between scales.
In summary, multiscale modeling requires at its foundation, measurement and communication of data between scales. To be useful to the biomedical engineer, the construction of quality computable grids and the one-way or two-way communication of data to and from these data over multiple scales must be fully automatic and accessible. One persistent challenge will be the registration of multiple datasets from different modalities to a common geometric database. While some of these concepts overlap with “atlasing” projects, such as the Allen Brain Atlas [62], there is a need to adapt these approaches to the needs of the modeling community, by developing procedures and algorithms that make the communication of data routine.
Priorities
Computational biomechanical modeling typically requires a level of customization beyond what is possible with commercial tools. This is even more forcefully true with multiscale modeling where codes that are specialized for different scales often must communicate – and more and more often in parallel. At the same time, a plethora of lab-specific codes entails unnecessary duplication, and commonly less effective codes due to the substantial investment required. In contrast, the open-source paradigm is a proven model for complex software development that has the capacity to effectively create computational tools that are geared to the needs of multiscale modeling, while enabling unlimited customization through access to the source code. A few examples of highly successful open-source projects of relevance to multiscale modeling are the image processing and registration toolkit, ITK [63], another similar tool, 3D Slicer [64], and the post-processor Paraview [65]. The NIH sponsored SimTK project [66] is another example that aims to bring together multiple components geared toward molecular, neuromuscular and cardiovascular dynamics. A few common aspects of successful open-source projects are that they are funded, supported by the biomedical community and well-organized. The organization of open-source efforts around task-oriented components that are designed to work together (image segmentation, grid generation and management, inverse parameter estimation, computational continuum mechanics, network models, system level models, ODE solvers, 1D and 2D PDE solvers, etc.) would greatly enhance the ability of multiscale modelers to focus on biomedical problem solving and discovery, the ultimate goals of biomedical multiscale modeling.
Model sharing also has the potential to empower the multiscale modeler by providing a library of models that can be combined across scales for a given application. One roadblock to effective model sharing, however, is the lack of a standard format. This problem has been recognized by computational practitioners outside of the biomedical field and has led to such projects as CGNS [67]. The goals of CGNS, as an example, provide an interesting model for the biomedical modeling community in that they provide, in collaboration with commercial entities, a standard format that is self-descriptive, machine-independent and well-documented. This paradigm has been so successful that virtually all major commercial computational fluid dynamics codes support it. However, CGNS is more than a format. It is a comprehensive, open-source data archiving system with standard naming conventions that provide definitions of boundary conditions, solver specifications and convergence history, such that a solution obtained in one commercial or research code can be restarted directly in another code. To assure consistent implementation the project provides easy-to-implement midlevel libraries in all major computer languages. Adoption of a common database and format is complimentary to a common interfaces approach and is foundational to successful open-source projects in multiscale modeling.
Dissemination of a solution database with model distribution may have practical value. Multiscale simulations involve many models at the bottom of the solution hierarchy, which usually have an input-output relationship with the models at a higher level. Solution for these models are requested frequently while solving for the higher level model and in many cases individual solutions are costly. This can be seen in multi-level finite element models of tissue-cell interactions [68] and in musculoskeletal modeling where rigid body movement simulations requesting simulations of a tissue level finite element model at each time step [41]. If a solution database exists, one can build a fast surrogate model to represent the input-output relationship. These surrogate representations can be based on a global fit (e.g., response surface [69], which were commonly used in optimization problems), local regression (e.g., moving least squares [70]), or neural networks [71]. For example, a surrogate type modeling approach uses discrete mass-spring-damper models (Fig. 3) to represent soft tissue. Parameters for the mass-spring-damper network are optimized to fit tissue deformations from either experimental measurements or finite element simulations [72]. Alternatively, adaptive strategies can be adopted, which estimate a fit or interpolation error and simulate the complicated model only when needed. By using adaptive surrogate modeling techniques based on local weighted learning [73] , it is now possible to solve optimal control problems that requires concurrent simulation of musculoskeletal movements and tissue deformations. Muscle activations for a jumping simulation (Fig. 1) can now be predicted by using a surrogate model of foot deformations. This approach requires costly finite element analysis to be conducted only for approximately 30% of the time.
Disclaimer
This article does not intend to be complete nor apply to all areas of multiscale modeling. The text is based on the discussions between the authors while sharing their experience to combine multiple physics, domains and scales in their computational biomechanics research.
Acknowledgment
Funding to support contributions of individual authors were: Ahmet Erdemir, National Institutes of Health, USA (1R01EB006735-01, Principal Investigator: Antonie J. van den Bogert); Daniel Einstein, National Institutes of Health, USA (1R01HL073598-01A1, Principal Investigator: Richard A. Corley); Trent Guess, National Science Foundation, USA, under the IMAG program for Multiscale Modeling (CMS-0506297); Merryn Tawhai, National Institutes of Health, USA, under the IMAG program for Multiscale Modeling (R01-EB-005823, Principal Investigator: Ching-Long Lin) and a Bioengineering Research Partnership Grant (R01-HL-064368, Principal Investigator: Eric A. Hoffman).
This article is written through collaborative editing using the Interagency Modeling and Analysis Group (IMAG) wiki [74]. The authors are grateful to IMAG for providing this platform and also appreciate the efforts of the developers of Mediawiki [75], a free wiki engine upon which this platform is based. An online version of this article in preprint form can be found in the IMAG wiki [76].
References
[1] K.G. Wilson, "Problems in physics with many scales of length," Scientific American, vol. 241, pp. 158–179, 1979.
[2] S.S. Blemker and S.L. Delp, "Rectus femoris and vastus intermedius fiber excursions predicted by three-dimensional muscle models," J. Biomech., vol. 39, pp. 1383-1391, 2006.
[3] A.D. Speirs, M.O. Heller, G.N. Duda, and W.R. Taylor, "Physiologically based boundary conditions in finite element modelling," J. Biomech., vol. 40, pp. 2318-2323, 2007.
[4] S.M. Cowan, P.W. Hodges, K.L. Bennell, and K.M. Crossley, "Altered vastii recruitment when people with patellofemoral pain syndrome complete a postural task," Arch. Phys. Med. Rehabil., vol. 83, pp. 989-995, 2002.
[5] E. Witvrouw, R. Lysens, J. Bellemans, D. Cambier, and G. Vanderstraeten, "Intrinsic risk factors for the development of anterior knee pain in an athletic population. A two-year prospective study," Am. J. Sports Med., vol. 28, pp. 480-489, 2000.
[6] E.M. Wojtys, D.N. Beaman, R.A. Glover, and D. Janda, "Innervation of the human knee joint by substance-P fibers," Arthroscopy, vol. 6, pp. 254-263, 1990.
[7] E. Panagiotopoulos, P. Strzelczyk, M. Herrmann, and G. Scuderi "Cadaveric study on static medial patellar stabilizers: the dynamizing role of the vastus medialis obliquus on medial patellofemoral ligament," Knee Surg. Sports Traumatol. Arthrosc., vol. 14, pp. 7-12, Jan. 2006.
[8] J.H. Koolstra, "Dynamics of the human masticatory system," Crit. Rev. Oral Biol. Med., vol. 13, pp. 366-376, 2002.
[9] L.Y. Griffin, J. Agel, M.J. Albohm, E.A. Arendt, R.W. Dick, W.E. Garrett, J.G. Garrick, T.E. Hewett, L. Huston, M.L. Ireland, R.J. Johnson, W.B. Kibler, S. Lephart, J.L. Lewis, T.N. Lindenfeld, B.R. Mandelbaum, P. Marchak, C.C. Teitz, and E.M. Wojtys, "Noncontact anterior cruciate ligament injuries: risk factors and prevention strategies," J. Am. Acad. Orthop. Surg., vol. 8, pp. 141-150, May-Jun. 2000.
[10] International Working Group on the Diabetic Foot, International consensus on the diabetic foot, International Diabetes Federation, 1999.
[11] S. Yagihashi, S. Yamagishi, and R. Wada, "Pathology and pathogenetic mechanisms of diabetic neuropathy: correlation with clinical signs and symptoms," Diabetes Res. Clin. Pract., vol 77, pp. S184-S189, 2007.
[12] O.Y. Kwon, S.D. Minor, K.S. Maluf, and M.J. Mueller, "Comparison of muscle activity during walking in subjects with and without diabetic neuropathy," Gait Posture, vol 18, pp. 105-113, Aug. 2003.
[13] R. Loganathan, M. Bilgen, B. Al-Hafez, and I.V. Smirnova IV, "Characterization of alterations in diabetic myocardial tissue using high resolution MRI," Int. J. Cardiovasc. Imaging', vol 22, pp. 81-90, Feb. 2006.
[14] M. Lorenzi and C. Gerhardinger, "Early cellular and molecular changes induced by diabetes in the retina," Diabetologia, vol. 44, pp. 791-804, Jul. 2001.
[15] S.A. Bus, Q.X. Yang, J.H. Wang, M.B. Smith, R. Wunderlich, and P.R. Cavanagh, "Intrinsic muscle atrophy and toe deformity in the diabetic neuropathic foot: a magnetic resonance imaging study," Diabetes Care vol. 25, pp. 1444-1450, Aug. 2002.
[16] J.J. Fredberg and R.D. Kamm, "Stress transmission in the lung: pathways from organ to molecule," Annu. Rev. Physiol., vol. 68, pp. 507-541, 2006.
[17] S.J. Lai-Fook, T.A. Wilson, R.E. Hyatt, and J.R. Rodarte, "Elastic constants of inflated lobes of dog lungs," Journal of Applied Physiology, vol. 40, pp. 508-513, 1976.
[18] J.M. Garcia-Aznar, J.H. Kuiper, M.J. Gomez-Benito, M. Doblare, and J.B. Richardson, "Computational simulation of fracture healing: Influence of interfragmentary movement on the callus growth," J. Biomech., vol. 40, pp. 1467-1476, 2007.
[19] J. Wolff, The Law of Bone Remodeling, Berlin: Springer-Verlag, 1986.
[20] G.F. Oster, J.D. Murray, and A.K. Harris, "Mechanical aspects of mesenchymal morphogenesis," J. Embryol. Exp. Morph., vol. 78, pp. 83-125, 1983.
[21] R. Huiskes, R. Ruimerman, G.H. van Lenthe, and J.D. Janssen, "Effects of mechanical forces on maintenance and adaptation of form in trabecular bone," Nature, vol. 405, pp. 704-706, 2000.
[22] S.J. Shefelbine, P. Augat, L. Claes, and U. Simon, "Trabecular bone fracture healing simulation with finite element analysis and fuzzy logic," J. Biomech., vol. 38, pp. 2440-2450, 2005.
[23] S.C. Cowin, "Tissue growth and remodeling," Ann. Rev. Biomed. Eng., vol. 6, pp. 77-107, 2004.
[24] R.L. Gleason and J.D. Humphrey, "Effects of a sustained extension on arterial growth and remodeling: a theoretical study," J. Biomech., vol. 38, pp. 1255-1261, 2005.
[25] K. Garikipati, J.E. Olberding, H. Narayanan, E.M. Arruda, K. Grosh, and S. Calve, "Biological remodelling: Stationary energy, configurational change, internal variables and dissipation," J. Mech. Phys. Solids, vol. 54, pp. 1493-1515, 2006.
[26] L.A. Taber and J.D. Humphrey, "Stress-modulated growth, residual stress, and vascular heterogeneity," J. Biomech. Eng., vol. 123, pp. 528-535, 2001.
[27] E. Kuhl, K. Garikipati, E.M. Arruda, and K. Grosh, "Remodeling of biological tissue: Mechanically induced reorientation of a transversely isotropic chain network," J. Mech. Phys. Solids, vol. 53, pp. 1552-1573, 2005.
[28] N.J.B. Driessen, G.W.M. Peters, J.M. Huyghe, C.V.C. Bouten, and F.P.T. Baaijens, "Remodeling of continuously distributed collagen fibers in soft connective tissues," J. Biomech., vol. 36, pp. 1151-1158, 2003.
[29] J.E. Bischoff, "Continuum approach for tissue remodeling mediated by matrix stiffness," presented at ASME Summer Bioeng. Conf., Amelia Island, FL, 2006.
[30] L.M. McNamara and P.J. Prendergast, "Bone remodelling algorithms incorporating both strain and microdamage stimuli," J. Biomech., vol. 40, pp. 1381-1391, 2007.
[31] V.A. Lubarda, "Constitutive theories based on the multiplicative decomposition of deformation gradient: Thermo elasticity, elastoplasticity, and biomechanics," Appl. Mech. Rev., vol. 57, pp. 95-108, 2004.
[32] V.H. Barocas and R.T. Tranquillo, "An anisotropic biphasic theory of tissue-equivalent mechanics: The interplay among cell traction, fibrillar network deformation, fibril alignment, and cell contact guidance," J. Biomech. Eng., vol. 119, pp. 137-145, 1997.
[33] D. Ambard and P. Swider, "A predictive mechano-biological model of the bone-implant healing," Eur. J. Mech. A/Solids, vol. 25, pp. 927-937, 2006.
[34] Y.-S.J. Li, J.H. Haga, and S. Chien, "Molecular basis of the effects of shear stress on vascular endothelial cells," J. Biomech., vol. 38, pp. 1949-1971, 2005.
[35] J.D. Humphrey, "Stress, strain, and mechanotransduction in cells," J. Biomech. Eng., vol. 123, pp. 638-641, 2001.
[36] T. F. Besier, G. E. Gold, G. S. Beaupre, and S. L. Delp, "A modeling framework to estimate patellofemoral joint cartilage stress in vivo," Med. Sci. Sports Exerc., vol. 37, pp. 1924-1930, Nov. 2005.
[37] F. Ezquerro, A. Simon, M. Prado, and A. Perez, "Combination of finite element modeling and optimization for the study of lumbar spine biomechanics considering the 3D thorax-pelvis orientation," Med. Eng. Phys., vol. 26, pp. 11-22, Jan. 2004.
[38] M.G. Pandy, "Computer modeling and simulation of human movement," Annu. Rev. Biomed. Eng., vol. 3, pp. 245-273, 2001.
[39] F.C. Anderson and M.G. Pandy, "A dynamic optimization solution for vertical jumping in three dimensions," Comput. Methods Biomech. Biomed. Engin. vol.2 , pp. 201-231, 1999.
[40] F.C. Anderson and M.G. Pandy, "Dynamic optimization of human walking," J. Biomech. Eng., vol. 123, pp. 381-390, 2001.
[41] A.J. van den Bogert, and A. Erdemir, "Concurrent simulations of musculoskeletal movements and tissue deformations,", presented at ASME Summer Bioengineering Conference, Keystone, CO, June 20-24, 2007.
[42] A. Erdemir, S. McLean, W. Herzog, and A. J. van den Bogert, "Model-based estimation of muscle forces exerted during movements," Clin. Biomech., vol. 22, pp. 131-154, Feb. 2007.
[43] T.A. Wilson, "Nonuniform lung deformations," J. Appl. Physiol., vol. 54, pp. 1443-1450, 1983.
[44] J.B. West, and F.L. Matthews, "Stresses, strains, and surface pressures in lung caused by its weight," J. Appl. Physiol., vol. 32, pp. 332-345, 1972.
[45] R. De Wilde, J. Clément, J.M. Hellemans, M. Decramer, M. Demedts, R. Boving, and K.P. Van de Woestijne, "Model of elasticity of the human lung," Journal of Applied Physiology, vol. 51, pp. 254-261, 1981.
[46] Y.C. Fung, Biomechanics: Motion, Flow, Stress, and Growth, 1st Ed., Springer, pp. 429.
[47] M.H. Tawhai, M.P. Nash and E.A. Hoffman, "An imaging-based computational approach to model ventilation distribution and soft-tissue deformation in the ovine lung," Acad. Radiol., vol. 13, pp. 113-120, Jan. 2006.
[48] H. Bachofen, P. Gehr, and E.R. Weibel, "Alterations of mechanical properties and morphology in excised rabbit lungs reinsed with a detergent," J. Appl. Physiol., vol. 47, pp. 1002-1010, 1979.
[49] T.A. Wilson and H. Bachofen, "A model of mechanical structure of alveolar duct," J. Appl. Physiol., vol. 52, pp. 1064-1070, 1982.
[50] E. Denny and R.C. Schroter, "A model of non-uniform lung parenchyma distortion," J. Biomech., vol. 39, pp. 652-663, 2006.
[51] S.L. Delp, F.C. Anderson, A.S. Arnold, P. Loan, A. Habib, C.T. John, E. Guendelman E, and D.G. Thelen, "OpenSim: open-source software to create and analyze dynamic simulations of movement," IEEE Transactions on Biomedical Engineering, vol. 54, pp. 1940-1950, Nov. 2007.
[52] http://mrl.sci.utah.edu/software.php, accessed on November 14, 2007.
[53] H. Si, "Adaptive tetrahedral mesh generation by constrained Delaunay refinement," Weierstrass Institute for Applied Analysis and Stochastics, Berlin, Germany, 2006.
[54] M. Sopics, "Taking on the ITER Challenge, Scientists Look to Innovative Algorithms, Petascale Computers," in SIAM News, vol. 39, 2006.
[55] A.P. Kuprat and D.R. Einstein, "An anisotropic scale-invariant unstructured tetrahedral mesh generation algorithm based on local feature size," Journal of Computational Physics, submitted, 2007.
[56] A.P. Kuprat and S.J. Mosso, "Volume conserving smoothing for piecewise linear curves, surfaces, and triple lines," Journal of Computational Physics, vol. 172, pp. 99-118, 2001.
[57] A.P. Kuprat and S.J. Mosso, "Efficient algortihms for mapping cell quantities between overlapped 3D unstructured meshes," Los Alamos National Laboratory Report, Los Alamos, NM, 2005.
[58] R.A. Burton, G. Plank, J.E. Schneider, V. Grau, H. Ahammer, S.L. Keeling, J. Lee, N.P. Smith, D. Gavaghan, N. Trayanova, and P. Kohl, "Three-dimensional models of individual cardiac histoanatomy: tools and challenges," Ann. N. Y. Acad. Sci. vol. 1080, pp. 301-319, Oct. 2006.
[59] T. Ju, J. Warren, J. Carson, M. Bello, I. Kakadiaris, W. Chiu, C. Thaller, and G. Eichele, "3D volume reconstruction of a mouse brain from histological sections using warp filtering," J. Neurosci. Methods., vol. 156, pp. 84-100, 2006.
[60] J.P. Carson, G. Eichele, and W. Chiu, "A method for automated detection of gene expression required for the establishment of a digital transcriptome-wide gene expression atlas," J. Microsc., vol. 217, pp. 275-281, 2005.
[61] J.P. Carson, T. Ju, H.C. Lu, C. Thaller, M. Xu, S.L. Pallas, M.C. Crair, J. Warren, W. Chiu, and G. Eichele, "A digital atlas to characterize the mouse brain transcriptome," PLoS Comput. Biol., vol. 1, pp. e41, 2005.
[62] http://www.brain-map.org, accessed on November 14, 2007.
[63] http://www.itk.org, accessed on November 13, 2007.
[64] http://www.slicer.org, accessed on November 13, 2007.
[65] http://www.paraview.org, accessed on November 13, 2007.
[66] http://www.simtk.org, accessed on November 13, 2007.
[67] http://www.cgns.org, accessed on November 13, 2007.
[68] R.G. Breuls, B.G. Sengers, C.W. Oomens, C.V. Bouten, and F.P. Baaijens, "Predicting local cell deformations in engineered tissue constructs: a multilevel finite element approach," J. Biomech. Eng., vol. 124, pp. 198-207, Apr. 2002.
[69] W.J. Roux, N. Stander, and R.T. Haftka, "Response surface approximations for structural optimization," Int. J. Numer. Meth. Engng., vol. 42, pp. 517-534, 1998.
[70] P. Lancaster and K. Salkauskas, "Surfaces generated by moving least squares methods," Mathematics of Computation vol. 37, pp. 141-158, 1981.
[71] I. García, J.D. Martín-Guerrero, E. Soria-Olivas, R.J. Martínez, S. Rueda, and R. Magdalena, "A neural network approach for real time collision detection.," in IEEE International Conference on Systems, Man and Cybernetics, 2002.
[72] M. Kia and T. M. Guess, “The Study of Menisci Effect in a Computational Knee Model," presented at the 6th Combined Meeting of the Orthopaedic Research Societies, Honolulu, HI, October 2007.
[73] C.G. Atkeson, A.W. Moore, and S. Schaal, "Locally weighted learning," Artificial Intelligence Review, vol. 11, pp. 11-73, 1997.
[74] https://www.imagwiki.nibib.nih.gov/, accessed on November 13, 2007.
[75] https://www.imagwiki.nibib.nih.gov/, accessed on November 13, 2007.
[76] https://www.imagwiki.nibib.nih.gov/content/ieee-embs-article/, accessed on April 27, 2008.
Figures
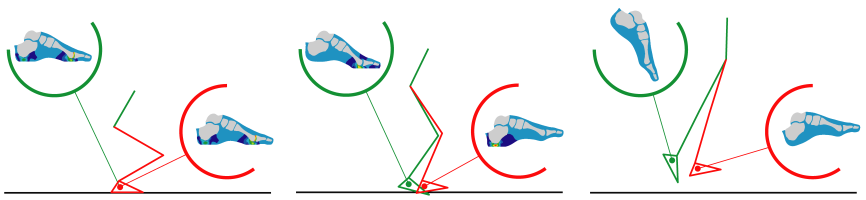
Figure 1. Concurrent simulation of multi-body dynamics and tissue deformations is possible as illustrated by this jumping simulation. Multibody dynamics was controlled by muscle actuation and calculated by forward dynamics solution of rigid body equations. Finite element analysis of the foot was conducted at each time step to predict foot stresses.
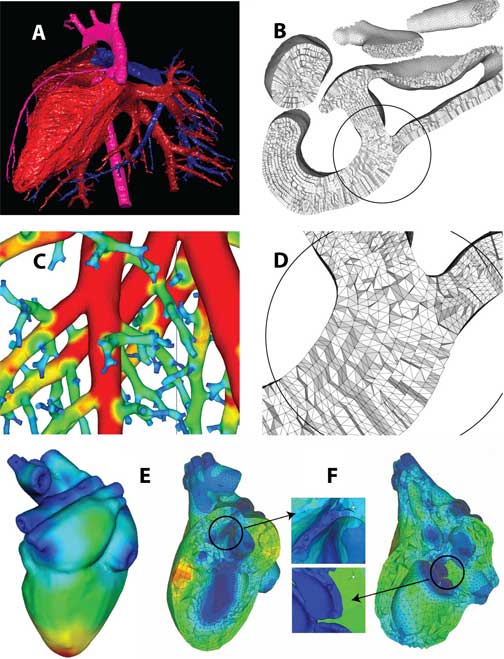
Figure 2. Automatic quality scale-invariant meshing: A) reconstruction of a mouse heart; B) computable grid of a mouse nose; C) feature-size field defined on a rat lung; D) detail of structured layered grid; E&F) feature-size field and structured mesh of a human heart (inset shows the mitral valve).
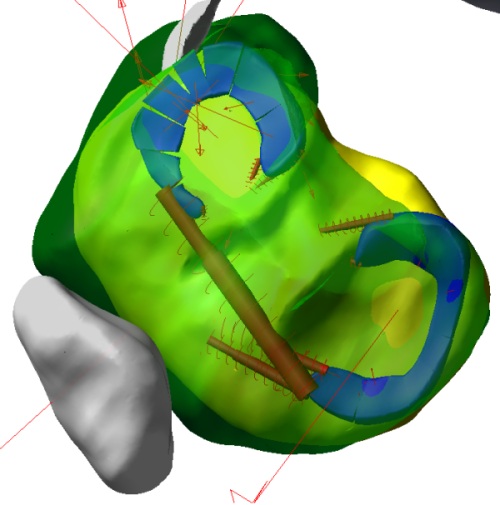
Figure 3. Discrete mass-spring-damper model of the human menisci during simulated walking. Model parameters were derived from optimization routines that minimized displacement error between simulations of a finite element meniscus model under identical loading.
Author Biographies

Merryn Tawhai received the Ph.D. in Engineering Science from the University of Auckland in 2001. Since then she has worked in the Auckland Bioengineering Institute at the University of Auckland, establishing a research team to develop integrative computational models of the lung for studying pulmonary physiology and pathophysiology. She also holds an adjunct appointment in the Department of Biomedical Engineering at the University of Iowa. Her main research interests have been in developing anatomically-based finite element models of physical structures in the lung, and using these individualized models to study lung tissue mechanics, ventilation distribution, pulmonary perfusion, airway thermofluid dynamics, and inert gas mixing.

Jeff Bischoff received his Ph.D. in Mechanical Engineering from the University of Michigan. He has held positions as a Lecturer in the Bioengineering Institute at the University of Auckland, Assistant Professor in Mechanical Engineering at the University of South Carolina, and is currently a Senior Research Engineer at Zimmer, Inc. His research interests include constitutive and computational modeling of soft tissue, orthopaedic biomechanics, multiscale modeling of growth and remodeling, experimental biomechanics, and material optimization. Current work involves development and implementation of novel inelastic constitutive models for soft materials including polymers and soft tissue, and use of computational techniques within joint reconstruction applications.

Daniel Einstein received his Ph.D. in bioengineering from the University of Washington. His work has included the development of computational methods for fluid-solid interactions, large deformation mechanics and inverse analysis. Since joining PNNL in 2005, he has established a small team of researchers to develop computational approaches for image-based multiscale modeling in the rat and mouse respiratory tract, and the application of these tools to understand the health effects of environmental exposure. Active collaborators who deserve mention are James P. Carson, Andrew P. Kuprat and Senthil Kabilan. Some of his active interests are developing novel approaches for biomedical grid generation, image segmentation and registration, nanoparticle mechanics, biological atlases, finite volume computational physics, and computational morphology.

Ahmet Erdemir, PhD, joined the Cleveland Clinic in 2002 following his graduate work at the Pennsylvania State University. He has training in biomechanics and mechanical engineering and he is particularly interested in musculoskeletal biomechanics and soft tissue mechanics at multiple scales. He has well-established research experience in computational biomechanics utilizing optimization techniques and finite element analysis. He also has expertise in cadaver experimentation and human subjects testing. He is an active reviewer for biomechanics related journals and holds memberships to many professional societies. Ahmet has established, and is currently directing, the Computational Biomodeling Core in the Department of Biomedical Engineering at the Cleveland Clinic to promote simulation-based medicine.

Trent Guess, Ph.D., is an Assistant Professor of Mechanical Engineering at the University of Missouri – Kansas City (UMKC) and holds an adjunct position in the Department of Orthopaedic Surgery in UMKC's School of Medicine. He is the director of the Musculoskeletal Biomechanics Research Lab at UMKC with research interests in the relationship between neuromuscular response and tissue loading. His research interests have led to the development of novel multiscale modeling techniques in musculoskeletal biomechanics. He is currently the lead of the IMAG multiscale modeling consortium tissue mechanics working group and is the advisor of UMKC’s human powered vehicle team.

Jeff Reinbolt, Ph.D., is a Distinguished Postdoctoral Fellow at Stanford University where he develops and applies software to study the dynamics and function of human health and disease. His appointment is within the Center for Physics-Based Simulation of Biological Structures, which is one of seven National Centers for Biomedical Computation supported by the National Institutes of Health Roadmap for Bioinformatics and Computational Biology. His research interests include: biomedical computation, simulation-based surgical and rehabilitation treatment planning, innovative patient-specific modeling, and optimization methods. He co-chaired a session at the Pacific Symposium on Biocomputing 2008 entitled Multiscale Modeling and Simulation: from Molecules to Cells to Organisms.

