We are exploring the possibility of creating a web-based resource where models relevant to viral pandemics can be hosted. The web site would allow the user to run models within their browser, including activities such as modifying model parameters. Since this Wiki does not have this capability, we are starting with a page that links to models at other sites where they can be run.
Models will hopefully cover a range of complexities, from simple SIR model in SBML running in an Jupyter Notebook (or perhaps as a NetLogo model) to complex multiscale models that include discreet cells, subcellular processes, immune response, etc.
Online Computational Platforms:
nanoHUB is funded by the NSF and others, and is an online computing host for a range of computational models in nanotechnology, physics and biology. NanoHUB supports a wide range of computing languages and has been used to tech both scientific and computing techniques. Running a model requires a user account, which is free. See https://nanohub.org/, to register visit https://nanohub.org/register/. Note that you must be a registered user and logged in to nanoHUB with your browser before the links to models given below will work properly. You can sign up for nanoHUB at https://nanohub.org/register/. For more info see:
-
Krishna Madhavan, et al. "nanoHUB.org: Cloud-based Services for Nanoscale Modeling, Simulation, and Education." Nanotechnology Reviews, Volume 2, Issue 1, Pages 107–117, ISSN (Online) 2191-9097, ISSN (Print) 2191-9089, DOI: 10.1515/ntrev-2012-0043, January 2013. https://www.degruyter.com/document/doi/10.1515/ntrev-2012-0043/html
-
Gerhard Klimeck, et al., “Network for Computational Nanotechnology – A Strategic Plan for Global Knowledge Transfer in Research and Education,” IEEE Nanotechnology Materials and Devices Conference (IEEE NMDC 2011), The Shilla Jeju, Korea, Oct. 18-21, 2011.
-
Gerhard Klimeck, et al., "nanoHUB.org: Advancing Education and Research in Nanotechnology," IEEE Computers in Engineering and Science (CISE), Vol. 10, pp. 17-23, 2008, doi:10.1109/MCSE.2008.120.
NetLogo (https://ccl.northwestern.edu/netlogo/) is is a modeling language for agent-based models (ABMs). It can be run as either a local application on your computer or remotely via a web browser. For more info see:
-
NetLogo itself: Wilensky, U. 1999. NetLogo. http://ccl.northwestern.edu/netlogo/. Center for Connected Learning and Computer-Based Modeling, Northwestern University. Evanston, IL.
-
HubNet: Wilensky, U. & Stroup, W., 1999. HubNet. http://ccl.northwestern.edu/netlogo/hubnet.html. Center for Connected Learning and Computer-Based Modeling, Northwestern University. Evanston, IL.
Model Repositories that don't include the ability to run models online:
BioModels: to retrieve all the immune and infection models available in BioModels try this url: https://www.ebi.ac.uk/biomodels/search?offset=0&numResults=10&sort=relevance-desc&query=infection+and+immune&domain=biomodels (returns 109 models as of July 10, 2023)
Runnable Models Index
- Sego et al., Multiscale spatial model of primary infection and immune response at nanoHUB (complex)
- MacKlin et al. COVID19 Tissue Simulator at nanoHUB (complex)
- Cockrell and An, Comparative Computational Modeling of the Bat and Human Immune Response to Viral Infection with the Comparative Biology Immune Agent Based Model (complex)
- Basic SIR model for viral spread in a population, part of the NetLogo's set of demos. (simple)
- Basic SIR model at nanoHUB. (simple)
Sego et al., A modular framework for multiscale, multicellular, spatiotemporal modeling of acute primary viral infection and immune response in epithelial tissues and its application to drug therapy timing and effectiveness
Reference: Sego TJ, Aponte-Serrano JO, Ferrari Gianlupi J, Heaps SR, Breithaupt K, Brusch L, Crawshaw J, Osborne JM, Quardokus EM, Plemper RK, Glazier JA. A modular framework for multiscale, multicellular, spatiotemporal modeling of acute primary viral infection and immune response in epithelial tissues and its application to drug therapy timing and effectiveness. PLoS Comput Biol. 2020 Dec 21;16(12):e1008451. doi: 10.1371/journal.pcbi.1008451. PMID: 33347439; PMCID: PMC7785254.
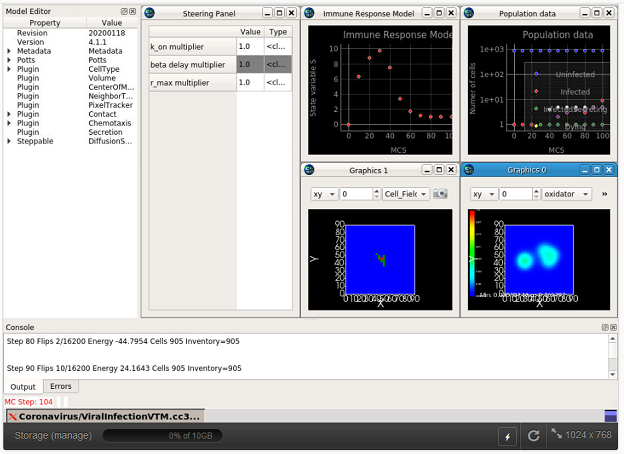
Model Location: This model can be run at the nanoHUB website (requires a nanoHUB account): https://nanohub.org/tools/cc3dcovid19. In addition, the entire project can be obtained from the github repository at https://github.com/covid-tissue-models/covid-tissue-response-models.
In support of parallel model development of tissue infection, we are developing a multiscale computational model of viral tissue infection and public repository of shared modeling and simulation developments. The repository will be maintained with standard documentation of shared models and simulation tools, for which we will provide supporting templates and generators.
What it does: Currently, the model describes select interactions between generalized epithelial and immune cells and their extracellular environment associated with viral infection and immune response at the cellular and subcellular levels. The repository contains the model and prototypical standard documentation.
How we built it: We are implementing all model components in CompuCell3D, an open-source modeling and simulation environment of the spatiotemporal dynamics of virtual tissue at the cellular and subcellular levels. Model components under development are written in python and simulated in the CompuCell3D simulation engine, which is developed in c++. Supporting tools for documentation standards are being developed in python using Sphinx.
MacKlin et al. COVID19 Tissue Simulator at nanoHUB
Reference: M. Getz et al., Rapid community-driven development of a SARS-CoV-2 tissue simulator. bioRxiv 2020.04.02.019075, 2020, (Preprint). doi: 10.1101/2020.04.02.019075.
This model simulates viral dynamics of SARS-CoV-2 (coronavirus / COVID19) in a layer of epithelium and several submodels (such as single-cell response, pyroptosis death model, tissue-damage model, lymph node model and immune response). It is being rapidly prototyped and refined with community support. This model includes a range of cell types and processes, for more details see here.
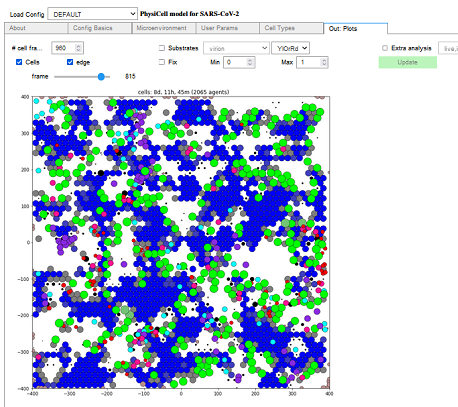
Model Location: This model can be run at the nanoHUB website (requires a nanoHUB account): https://nanohub.org/resources/pc4covid19
What it does: tbd
How we built it: tbd
Cockrell and An, Comparative Computational Modeling of the Bat and Human Immune Response to Viral Infection with the Comparative Biology Immune Agent Based Model
Reference: Cockrell C, An G. Comparative Computational Modeling of the Bat and Human Immune Response to Viral Infection with the Comparative Biology Immune Agent Based Model. Viruses. 2021 Aug 16;13(8):1620. doi: 10.3390/v13081620. PMID: 34452484; PMCID: PMC8402910.
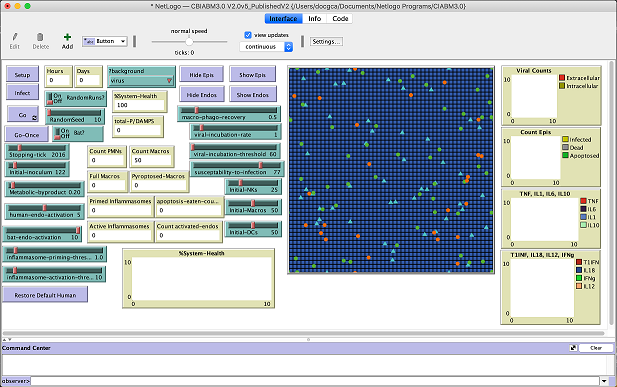
Model Location: This model can be run online at NetLogo using https://netlogoweb.org/web?https://raw.githubusercontent.com/An-Cockrell/Comparative-Biology-Immune-ABM/main/CBIABM3.0%20V2.0v5_PublishedV2. To do a quick run, click the buttons in the upper left "Setup", "Infect" then "Go". Scroll down and click the "Model Info" tab to get a brief description of the model or visit the reference link above. For help on using the NetLogo model see https://www.mdpi.com/article/10.3390/v13081620/s1. In addition, the entire project can be obtained from the GitHub repository at https://github.com/An-Cockrell/Comparative-Biology-Immune-ABM.
What it does: This agent based model examines differences in innate immune mechanisms between bats and humans. This model is largely focused on differences in inflammatory response in bats versus humans. Simulation experiments demonstrate the efficacy of bat-related features in conferring viral tolerance and also suggest a crucial role for endothelial inflammasome activity as a possible explanation of the diffidence in viral tolerance between bats and humans.
How we built it: This model is implemented in NetLogo (https://ccl.northwestern.edu/netlogo/).
Basic SIR model for viral spread in a population, part of the NetLogo's set of demos. (simple)
Reference:
- For the model itself: Wilensky, U. (1998). NetLogo Virus model. http://ccl.northwestern.edu/netlogo/models/Virus. Center for Connected Learning and Computer-Based Modeling, Northwestern University, Evanston, IL.NetLogo software as: Wilensky, U. (1999).
- NetLogo. http://ccl.northwestern.edu/netlogo/. Center for Connected Learning and Computer-Based Modeling, Northwestern University, Evanston, IL.
WHAT IS IT?: This model simulates the transmission and perpetuation of a virus in a human population.
Ecological biologists have suggested a number of factors which may influence the survival of a directly transmitted virus within a population. (Yorke, et al. "Seasonality and the requirements for perpetuation and eradication of viruses in populations." Journal of Epidemiology, volume 109, pages 103-123)
HOW IT WORKS: The model is initialized with 150 people, of which 10 are infected. People move randomly about the world in one of three states: healthy but susceptible to infection (green), sick and infectious (red), and healthy and immune (gray). People may die of infection or old age. When the population dips below the environment's "carrying capacity" (set at 300 in this model) healthy people may produce healthy (but susceptible) offspring. For information and descriptions of the other parameters in the model see https://ccl.northwestern.edu/netlogo/models/Virus.
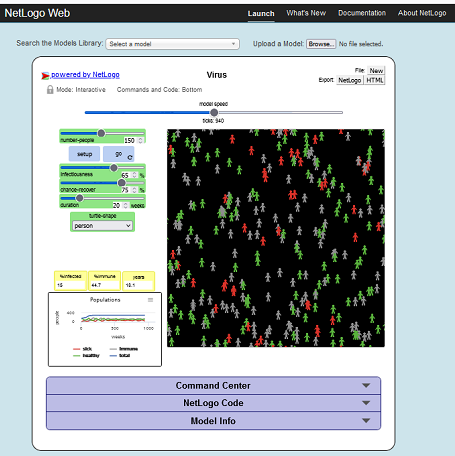
Model Location: This model can be run online at NetLogo at http://www.netlogoweb.org/launch#http://ccl.northwestern.edu/netlogo/models/models/Sample%20Models/Biology/Virus.nlogo
Basic SIR model in a Jupyter Notebook at nanoHUB (simple)
Reference:
Josua Oscar Aponte-Serrano (2020), "COVID-19 R0 Estimator," https://nanohub.org/resources/r0estimator2. (DOI: 10.21981/8Z8N-JD89).
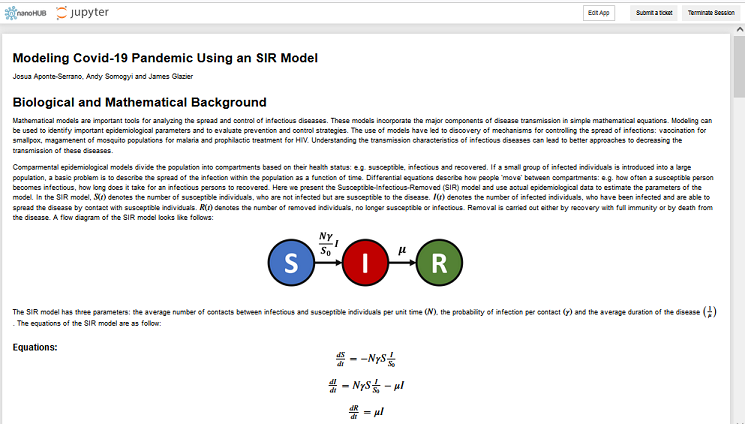
WHAT IS IT?: (From the link) Mathematical models are important tools for analyzing the spread and control of infectious diseases. These models incorporate the major components of disease transmission in simple mathematical equations. Modeling can be used to identify important epidemiological parameters and to evaluate prevention and control strategies. The use of models have led to discovery of mechanisms for controlling the spread of infections: vaccination for smallpox, magamenent of mosquito populations for malaria and prophilactic treatment for HIV. Understanding the transmission characteristics of infectious diseases can lead to better approaches to decreasing the transmission of these diseases.
Comparmental epidemiological models divide the population into compartments based on their health status: e.g. susceptible, infectious and recovered. If a small group of infected individuals is introduced into a large population, a basic problem is to describe the spread of the infection within the population as a function of time. Differential equations describe how people 'move' between compartments: e.g. how often a susceptible person becomes infectious, how long does it take for an infectious persons to recovered. Here we present the Susceptible-Infectious-Removed (SIR) model and use actual epidemiological data to estimate the parameters of the model.
HOW IT WORKS: See the detailed documentation inside the Jupyter notebook.
Model Location: This model can be run online at nanoHUB using https://nanohub.org/tools/r0estimator2. You will need an account on nanoHUB (https://nanohub.org/register/). After logging in, use the hyperlink above and then click on the "Launch Tool" button.
Disclaimer
Substantial effort has been made to provide accurate and complete information on this Web site. However, we cannot guarantee that there will be no errors. Neither the agencies of Interagency Modeling and Analysis Group (IMAG) nor the members of the Multiscale Modeling (MSM) Consortium assumes any legal liability for the accuracy, completeness, or usefulness of any information, products, or processes disclosed herein, or represents that use of such information, products, or processes would not infringe on privately owned rights. The views and opinions of authors expressed on the IMAG/MSM website does not necessarily state or reflect those of the IMAG or MSM Consortium, and they may not be used for advertising or product endorsement purposes.

