This document describes some basics techniques for using ordinary differential equations (ODEs) within MML models.
Prerequisites:
- Introductory Survey of MML (required)
- Introduction to the JSim GUI (recommended)
Contents:
- First example
- Systems of ODEs
- Second order equations
- Using implicit equations with ODEs
- Parameterized parallel systems
- Parameterized serial systems
- Using events with ODEs
- Comments or Questions?
First example
MML uses the colon (:) to represent differentiation (for example u:t represents du/dt). This construct is used in formulating ODEs in MML. Consider the following model of radioactive decay:
// radioactive decay
unit conversion on;
import nsrunit;
math main {
realDomain t sec;
t.min=0; t.max=4; t.delta=0.1;
real rate = 1 1/sec;
real u(t) kg; // ODE variable declaration
when (t=t.min) u=1; // initial condition for u
u:t = -rate*u; // state equation for u
}
JSim requires an ODE variable (here "u") be given an "initial condition (IC)" and a "state equation". The IC constrains the initial value by applying the "when" clause. In the example, the simplest possible constraint is used, that is, setting the numeric value. In more complicated systems, ICs may be calculated from other model variables. The state equation constrains the variable's first derivative (here "u:t") for all time. (For simplicity, the word "time" is used in this document to refer to an ODE variable's domain, however ODE variable domains be assigned any unit appropriate to the model.)
If you run the above model in the JSim GUI, you will discover that JSim creates a new IC variable u__init with default value 1 (matching the MML equation). This allows you to change the IC in the GUI without recompiling the model. If the IC for an ODE variable is calculated from other variables, the initial value will not be directly changable within the GUI.
Note that IC's when clause must be formulated as "t=t.min". You may not use "t=0" or any other specific numeric value in an IC.
Systems of ODEs
Consider the following model of diffusion between 3 compartments:
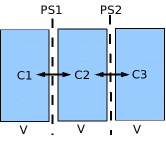
// 3 compartment diffusion
unit conversion on;
import nsrunit;
math comp3 {
realDomain t sec; // time
t.min=0; t.max=30; t.delta=0.2;
real V=.07 ml; // compartment volumes
real PS1=.05 ml/sec; // perm * surf area between compartments 1 & 2
real PS2=.02 ml/sec; // perm * surf area between compartments 2 & 3
real C1(t) mM, C2(t) mM, C3(t) mM; // conc in compartments 1,2,3
when (t=t.min) { C1=5; C2=3; C3=0; } // initial concentrations
C1:t = PS1/V*(C2 - C1); // C1 state eqn
C2:t = PS1/V*(C1 - C2) + PS2/V*(C3-C2); // C2 state eqn
C3:t = PS2/V*(C2 - C3); // C3 state eqn
}
Note that each of the three ODE variables (C1, C2, C3) has both an initial condition (specified via the "when" clause) and a state equation. The number of such equation in a JSim model is limited only by computer memory, not by JSim itself.
Second order equations
2nd (and higher) order ODEs may be solved in JSim. A Nth order ODE require N initial conditions (for u, u:t, ...). The following example that generates a sinusoid via a 2nd order ODE:
// 2nd order ODE: u:t:t = -u
unit conversion on;
import nsrunit;
math order2 {
realDomain t sec; // time
t.min=0; t.max=30; t.delta=0.2;
real u(t) m; //
when (t=t.min) {
u = 0;
u:t = 1;
}
u:t:t = (-1 1/sec^2) * u;
}
Using implicit equations with ODEs
You may use linear or non-linear implicit equations to specify ODE ICs or state equations. Consider the following example using linear implicit equations to specify both ICs and state equations:
// ODEs with implicit IC and state equations
unit conversion on;
import nsrunit;
math ode_implicit {
realDomain t sec;
t.min=0; t.max=10; t.delta=0.1;
real u(t) m, v(t) m;
when (t=t.min) {
u+v = 3;
u-v = 1;
}
u:t + v:t = (3 m/sec^2) * t;
u:t - 2*v:t = 0;
}
Here JSim will solve u and v at t=t.min via the two implicit equations in the when clause, and solve u:t and v:t via the last two equations. There are a number of tricky issues regarding implicit equations in MML. Model writers are strongly advised to read Implicit Equations in MML completely before using this construct in their own models.
Parameterized parallel systems
In some cases, you may wish to create a set of ODE variables that share a common form. The following code models flow through N parallel compartments:
// flow through parallel compartments
unit conversion on;
import nsrunit;
math comp3 {
realDomain t sec; // time
t.min=0; t.max=30; t.delta=0.2;
real N=5; // # compartments
realDomain n; // index of parallel compartments
n.min=1; n.max=N; n.delta=1; // n values slaved to N
private n.min, n.max, n.delta; // prevent user alterations
real F = .1 ml/sec; // flow rate
real V(n)=n * (.07 ml); // compartment volumes
real Cin(t) = (5 mM) * sin(t/(1 sec)); // input concentration
real C(t, n) mM; // conc in compartments 1-N
when (t=t.min) C = 0; // initial concentrations
C:t = F/V*(Cin - C); // state equation(s)
real Cout(t) = sum((C*V)@n)/sum(V@n); // output concentration
}
Here a realDomain "n" is introduced to parameterize the N parallel variables. N is initially set to 5, but may be easily changed in the JSim GUI. The ODE variable C now takes both time and "n" as domains, and the IC and state equations apply to each value of "n". This approach make for more concise and efficient code when a set of variables share a common equation form, differing only in parameter values. In this case, compartment volume (V) varies over "n".
Special notes:
- The combined output concentration is calculated via the MML sum() operator. See Using Integrals and Summations in MML for details on summations.
- The "private" statement ensures that N is the only variable affecting the values of "n". See Public, Private and Extern variables for more information.
- JSim internals issues currently prevent the value of N being set to 1. This limitation will be addressed in future JSim releases.
Parameterized serial system
The following code models flow through a series of N compartments:
// flow through serial compartments
unit conversion on;
import nsrunit;
math comp3 {
realDomain t sec; // time
t.min=0; t.max=30; t.delta=0.2;
real N=5; // # compartments
realDomain n; // index of parallel compartments
n.min=1; n.max=N; n.delta=1; // n values slaved to N
private n.min, n.max, n.delta; // prevent user alterations
real F = .1 ml/sec; // flow rate
real V(n)=n * (.07 ml); // compartment volumes
real Cin(t) = (5 mM) * sin(t/(1 sec)); // input concentration
real C(t, n) mM; // conc in compartments 1-N
when (t=t.min) C = 0; // initial concentrations
C:t = F/V*(if (n=1) Cin-C else C(t,n-1)-C); // state equation(s)
real Cout(t); // output concentration
when (n=n.max) Cout=C; // Cout is value for C at n=n.max
}
Most of the code is similar to the parallel example. The state equation uses MML's "if/else" construct to compare a compartment's concentration to its predecessor's. The 1st compartment is compared to the input (Cin).
The final "when" statement calculates Cout from the value of C at maximum n. This "when" construct is preferred to following which, although possible, is not recommended due to technical reason:
Cout=C(t,N);
Using events with ODEs
An MML ODE variable is inherently continuous, since it is calculated from the integral of a real-valued function. Certain physiological systems, such as gate openings and closings, are inherently discontinuous. Events are MML's construct for such discrete changes. See Using Events in MML for complete details.
Comments or Questions?
Model development and archiving support at https://www.imagwiki.nibib.nih.gov/physiome provided by the following grants: NIH U01HL122199 Analyzing the Cardiac Power Grid, 09/15/2015 - 05/31/2020, NIH/NIBIB BE08407 Software Integration, JSim and SBW 6/1/09-5/31/13; NIH/NHLBI T15 HL88516-01 Modeling for Heart, Lung and Blood: From Cell to Organ, 4/1/07-3/31/11; NSF BES-0506477 Adaptive Multi-Scale Model Simulation, 8/15/05-7/31/08; NIH/NHLBI R01 HL073598 Core 3: 3D Imaging and Computer Modeling of the Respiratory Tract, 9/1/04-8/31/09; as well as prior support from NIH/NCRR P41 RR01243 Simulation Resource in Circulatory Mass Transport and Exchange, 12/1/1980-11/30/01 and NIH/NIBIB R01 EB001973 JSim: A Simulation Analysis Platform, 3/1/02-2/28/07.

