This Gentex-based model simulates the transport and metabolism processes of exogenous Xa in the isolated, perfused non-working guinea pig heart, based on data from the multiple-indicator dilution technqiue (MID).
Description
This model simulates the transport and metabolism processes of exogenous Xanthine (Xa) and its metabolite utric acid in the isolated, perfused non-working guinea pig heart, based on the data acquired with the multiple-indicator dilution technqiue under the normoxia condition [Schwartz 1999]. The model were developed based on a gentric tissues exchange model,Gentex [Bassingthwaight 2006], which is a whole organ model of the vascular network providing intraorgan flow heterogeneity and accounts for substrate transmembrane transport, binding, and metabolism in erythrocytes, plasma, endothelial cells, interstitial space, and cardiomyocytes.
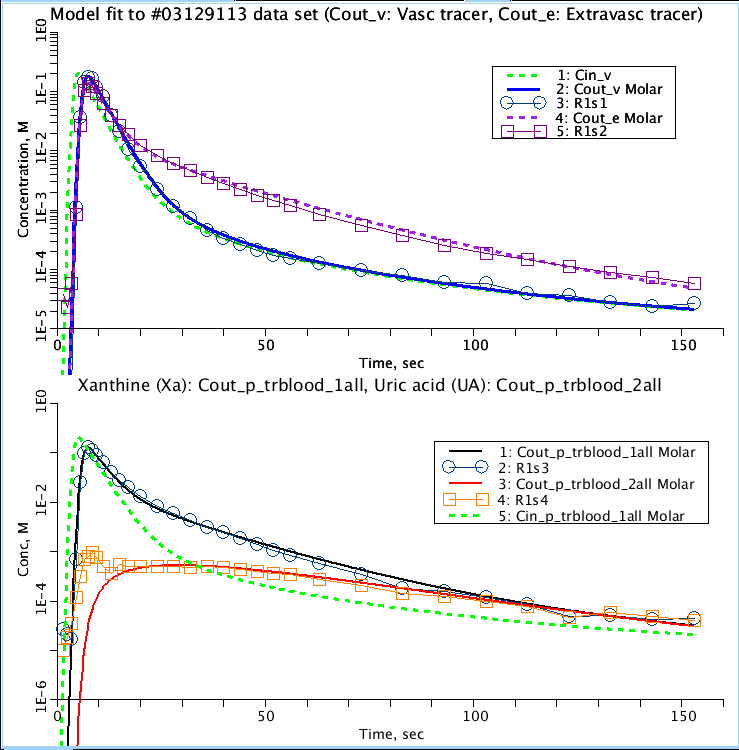
Figure: Top: Model fit to the dilution curve for the intravascular reference tracer, Albumin (blue), and extravascular reference tracer, L-Glucose (purple). Input function (green), assume a Lag-normal distribution, was derived from curve fitting the dilution curve for the intravascular reference. Bottom: Model fit for the dilution curve of injected tracer Xa (black) and its metabolite UA (red).
Equations
The equations for this model may be viewed by running the JSim model applet and clicking on the Source tab at the bottom left of JSim's Run Time graphical user interface. The equations are written in JSim's Mathematical Modeling Language (MML). See the Introduction to MML and the MML Reference Manual. Additional documentation for MML can be found by using the search option at the Physiome home page.
- Download JSim model MML code (text):
- Download translated SBML version of model (if available):
- No SBML translation currently available.
- Information on SBML conversion in JSim
We welcome comments and feedback for this model. Please use the button below to send comments:
Poulain CA, Finlayson BA, Bassingthwaighte JB.,Efficient numerical methods for nonlinear-facilitated transport and exchange in a blood-tissue exchange unit, Ann Biomed Eng. 1997 May-Jun;25(3):547-64. Bassingthwaighte JB, Raymond GR, Ploger JD, Schwartz LM, and Bukowski TR. GENTEX, a general multiscale model for in vivo tissue exchanges and intraorgan metabolism. Phil Trans Roy Soc : Mathematical, Physical and Engineering Sciences 2006. Schwartz LM, Bukowski TR, Revkin J adn Bassingthwaighte JB. Cardiac endothelial transport and metabolism of adenosine. Am J Physiol. 1999; 277(3): H1241-H1251. Kroll K, Bukowski TR, Schwartz LM, Knoepfler D, and Bassingthwaighte JB. Capillary endothelial transport of uric acid in guinea pig heart. Am. J. Physiol. 262(31): H420-H431. King RB, Bassingthwaighte JB, Hales JRS, Rowell LV. Stability of heterogeneity of myocardial blood flow in normal awake baboons. Circ. Res. 1985, 57:285-295.
Please cite https://www.imagwiki.nibib.nih.gov/physiome in any publication for which this software is used and send one reprint to the address given below:
The National Simulation Resource, Director J. B. Bassingthwaighte, Department of Bioengineering, University of Washington, Seattle WA 98195-5061.
Model development and archiving support at https://www.imagwiki.nibib.nih.gov/physiome provided by the following grants: NIH U01HL122199 Analyzing the Cardiac Power Grid, 09/15/2015 - 05/31/2020, NIH/NIBIB BE08407 Software Integration, JSim and SBW 6/1/09-5/31/13; NIH/NHLBI T15 HL88516-01 Modeling for Heart, Lung and Blood: From Cell to Organ, 4/1/07-3/31/11; NSF BES-0506477 Adaptive Multi-Scale Model Simulation, 8/15/05-7/31/08; NIH/NHLBI R01 HL073598 Core 3: 3D Imaging and Computer Modeling of the Respiratory Tract, 9/1/04-8/31/09; as well as prior support from NIH/NCRR P41 RR01243 Simulation Resource in Circulatory Mass Transport and Exchange, 12/1/1980-11/30/01 and NIH/NIBIB R01 EB001973 JSim: A Simulation Analysis Platform, 3/1/02-2/28/07.

