Four region blood-tissue exchange (BTEX) model used to fit data from Schwartz 1999 MID experiments involving cardiac endothelial transport and metabolism of adenosine and inosine.
Description
This blood-tissue exchange (BTEX) model represents a "tissue cylinder" consisting of four regions. The four regions are capillary plasma, p; endothelial cell, ec; interstitial fluid, isf; and parenchymal cell, pc; and are separated by three barriers--the luminal or plasma surface and endothelial cell layer; the albuminal surface of the endothelial cell facing the interstitium; andthe membrane between the interstitial fluid and parenchymal cell. In addition, there is a diffusional path from plasma to ISF bypassing endothelial cells via intercellular clefts. The model is used to fit data from Adenosine and Inosine uptake and metabolism experiments in animal hearts done by Schwartz et al. 1999. Metabolism within a region is modeled as constant or first-order consumption term.
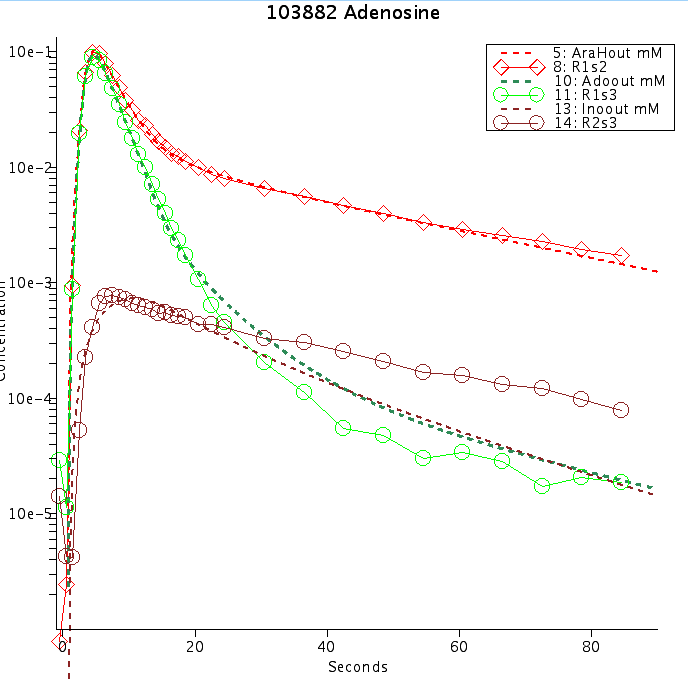
Figure: Adenosine (Ado -green) transport and metabolism in rabbit heart as a function of time after injection. Red is AraH, an extravascular reference, and brown is Inosine (Ino) a metabolite of Ado. Dashed lines are model fit to data (Schwartz 1999). Concentration is log scale.
Equations
The equations for this model may be viewed by running the JSim model applet and clicking on the Source tab at the bottom left of JSim's Run Time graphical user interface. The equations are written in JSim's Mathematical Modeling Language (MML). See the Introduction to MML and the MML Reference Manual. Additional documentation for MML can be found by using the search option at the Physiome home page.
- Download JSim model MML code (text):
- Download translated SBML version of model (if available):
- No SBML translation currently available.
- Information on SBML conversion in JSim
We welcome comments and feedback for this model. Please use the button below to send comments:
- Schwartz L. M., Bukowski T. R., Revkin J. H., Bassingthwaighte J. B.; Cardiac endothelial transport and metabolism of adenosine and inosine, AJP. Heart Circ. Physiol., 277, (H1241-H1251}, 1999
- Schwartz L. M., Bukowski T. R., Ploger J. D.; Bassingthwaighte J. B., Endothelial adenosine transporter characterization in perfused guinea pig hearts, AJP. Heart Circ. Physiol., 279, (H1502-H1511}, 2000
- Berne, Robert M; The Role of Adensoine in the Regulation of Coronary Blood Flow, Circulation Research, Vol 47:807-813, 1980
Please cite https://www.imagwiki.nibib.nih.gov/physiome in any publication for which this software is used and send one reprint to the address given below:
The National Simulation Resource, Director J. B. Bassingthwaighte, Department of Bioengineering, University of Washington, Seattle WA 98195-5061.
Model development and archiving support at https://www.imagwiki.nibib.nih.gov/physiome provided by the following grants: NIH U01HL122199 Analyzing the Cardiac Power Grid, 09/15/2015 - 05/31/2020, NIH/NIBIB BE08407 Software Integration, JSim and SBW 6/1/09-5/31/13; NIH/NHLBI T15 HL88516-01 Modeling for Heart, Lung and Blood: From Cell to Organ, 4/1/07-3/31/11; NSF BES-0506477 Adaptive Multi-Scale Model Simulation, 8/15/05-7/31/08; NIH/NHLBI R01 HL073598 Core 3: 3D Imaging and Computer Modeling of the Respiratory Tract, 9/1/04-8/31/09; as well as prior support from NIH/NCRR P41 RR01243 Simulation Resource in Circulatory Mass Transport and Exchange, 12/1/1980-11/30/01 and NIH/NIBIB R01 EB001973 JSim: A Simulation Analysis Platform, 3/1/02-2/28/07.

