Tracer and Mother solute flow past a binding site. Equilibrium upset by an added pulse of mother solute, dislodging tracer.
Description
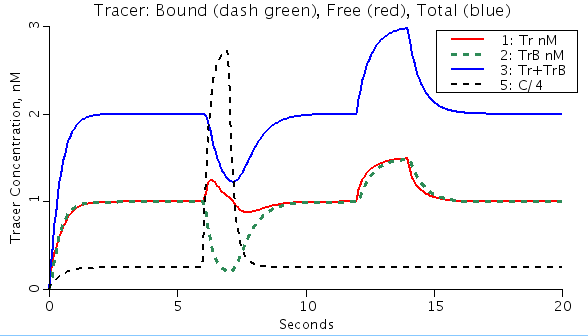
One compartment model of mother solute C and tracer labeled C, called Tr, with slow binding to a binding ligand B os constant total, Btotal, and which does not leave the flowing region..Volume V is constant. Total concentration of binding ligand, Btotal = unbound B and the complex CB. Tracer and mother substance flow in and out. Concn of unbound ligand is determined by the concns of mother and tracer analytes relative to the Kd mM, which is the dissociation constant for ligand-solute binding. The purpose of this model is to demonstrate that when the concn of mother substance is raised, tracer that has been attached to a binding ligand is driven off and washed out. The function generator fgen_1 is set up to provide a constant infusion of mother substance at 1 mM to which an additional 1-second pulse is added at t = 6 seconds. The tracer infusion is constant at 1 nM through to 12 seconds. The concentrations and the Kd are chosen ao that the pulse of mother solute suddenly reduces the amount of free ligand, and competes with the tracer that is already attached, causing a transient release of tracer and a sudden diminution in the total amount of tracer present in the mixing chamber. An external detector would give a signal proportional to Tr + TrB, the total tracer present. At 12 seconds a 2 second pulse of additional tracer is added, but if at a tracer level it has no effect on the mother substance binding. Explore the influence of making this pulse large by changing "p2amplitude" for Pulse 2 in fgen_2. Constants: Btotal = total concentration of binding ligand, mmol/ml V = Volume of compartment, ml kon = binding rate, 1/mmol*sec koff = off rate, 1/sec Variables: C(t) = Concentration of mother analyte, mmol/ml Tr(t) = Concentration of tracer analyte, nmol/ml CB(t) = Concentration of bound mother analyte with ligand B, mmol/ml TrB(t) = Concentration of bound tracer with ligand B, nmol/ml free Ligand [B] = [Btotal]- [CB]-[TrB]
Equations
The equations for this model may be viewed by running the JSim model applet and clicking on the Source tab at the bottom left of JSim's Run Time graphical user interface. The equations are written in JSim's Mathematical Modeling Language (MML). See the Introduction to MML and the MML Reference Manual. Additional documentation for MML can be found by using the search option at the Physiome home page.
- Download JSim model MML code (text):
- Download translated SBML version of model (if available):
We welcome comments and feedback for this model. Please use the button below to send comments:
Bassingthwaighte JB and Goresky CA. Modeling in the analysis of solute and water exchange in the microvasculature. In: Handbook of Physiology. Sect. 2, The Cardiovascular System. Vol IV, The Microcirculation, edited by Renkin EM and Michel CC. Bethesda, MD: Am. Physiol. Soc., 1984, pp 549-626. ( See Permeation p.560.)
Please cite https://www.imagwiki.nibib.nih.gov/physiome in any publication for which this software is used and send one reprint to the address given below:
The National Simulation Resource, Director J. B. Bassingthwaighte, Department of Bioengineering, University of Washington, Seattle WA 98195-5061.
Model development and archiving support at https://www.imagwiki.nibib.nih.gov/physiome provided by the following grants: NIH U01HL122199 Analyzing the Cardiac Power Grid, 09/15/2015 - 05/31/2020, NIH/NIBIB BE08407 Software Integration, JSim and SBW 6/1/09-5/31/13; NIH/NHLBI T15 HL88516-01 Modeling for Heart, Lung and Blood: From Cell to Organ, 4/1/07-3/31/11; NSF BES-0506477 Adaptive Multi-Scale Model Simulation, 8/15/05-7/31/08; NIH/NHLBI R01 HL073598 Core 3: 3D Imaging and Computer Modeling of the Respiratory Tract, 9/1/04-8/31/09; as well as prior support from NIH/NCRR P41 RR01243 Simulation Resource in Circulatory Mass Transport and Exchange, 12/1/1980-11/30/01 and NIH/NIBIB R01 EB001973 JSim: A Simulation Analysis Platform, 3/1/02-2/28/07.

