An enzyme E is synthesized at a constant rate, and degraded in a first order process, -ke*E, only when it is unbound. Input of S is sinusoidal. The system amplifies to give an oscillation in product P. From Reich and Selkov 1981 and really from Selkov 1968.
Description
Grisolia (1964) noted that enzyme degradation was a normal and apparently regulated process, and that the rates of degradation were much higher when the enzyme was not bound to substrate. The system for cleaning out damaged proteins takes place mostly in lyzosomes where the proteins are taken apart and the amino acids made available for new protein building. The process, autophagy, or eating of self, was identified by Ciechanover (2012 for his review) as an important feature of cell survival, keeping the house clean and tidy, for which he was awarded a Nobel Prize. In mice, He and colleagues found that rates of autophagy were increased by exercise, and that the mice lived longer. The model, taken from Reich and Sel'kov (1981), assumes a constant rate of synthesis, vE, of the enzyme E by transcription from the genome, and lysis of E by a first order hydrolysis, kE. Substrate S is supplied at a sinusoidally varying rate Sinp. Under the conditions, the enzyme is almost fully bound most of the time, i.e. ES/(ES+E) is close to 1.0 so there is little degradataion. When S gets used up, ES -> E and degradation occurs.
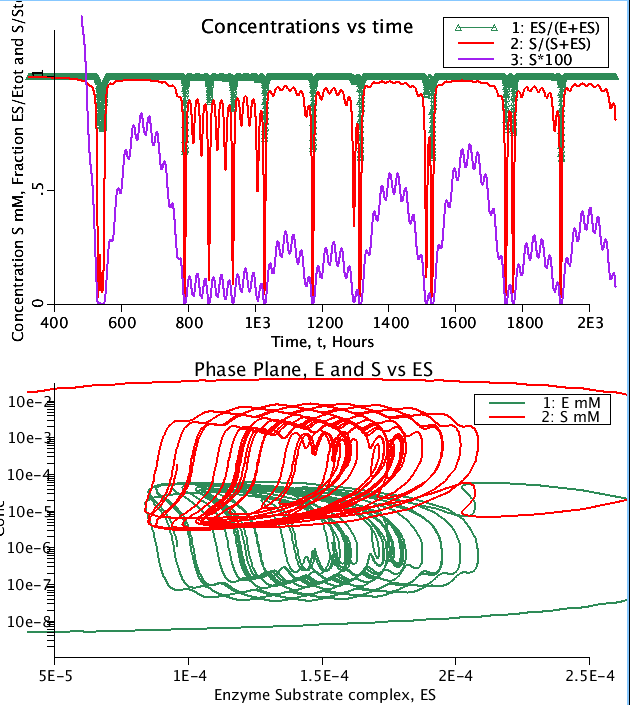
Figure: (Top) Concentration time course. (Bottom) Phase plane diagram of Concentration of E and S versus enzyme substrate complex (ES) concentration. Simulation run with low equilbrium dissociation constant (keq = 1e-8 mM)
Equations
The equations for this model may be viewed by running the JSim model applet and clicking on the Source tab at the bottom left of JSim's Run Time graphical user interface. The equations are written in JSim's Mathematical Modeling Language (MML). See the Introduction to MML and the MML Reference Manual. Additional documentation for MML can be found by using the search option at the Physiome home page.
- Download JSim model MML code (text):
- Download translated SBML version of model (if available):
We welcome comments and feedback for this model. Please use the button below to send comments:
Ciechanover A. Intracellular Protein Degradation: From a Vague Idea through the Lysosome and the Ubiquitin- Proteasome System and onto Human Diseases and Drug Targeting. Ramban Maimonides Med J 3: 1-20, 2012. He C, Bassik MC, Moresi V, Sun K, Wei Y, Zou Z, An Z, Loh J, Fisher J, Sun Q, Korsmeyer S, Packer M, May HI, Hill JA, Virgin HW, Gilpin C, Xiao G, Bassel-Duby R, Scherer PE, and Levine B. Exercise-induced BCL2-regulated autophagy is required for muscle glucose homeostasis. Nature advance online publication: 2012/01/18/online, 2012/01/18/online. Grisolia S. The catalytic environment and its biological implications. Physiol Rev 44: 657-714, 1964. Reich JG and Sel'kov EE. Energy Metabolism of the Cell: A Theoretical Treatise. London: Academic Press, 1981. (p 40) Selkov EE. Self-oscillations in glycolysis. 1. A simple kinetic model. European J Biochem 4: 79-86, 1968.
Please cite https://www.imagwiki.nibib.nih.gov/physiome in any publication for which this software is used and send one reprint to the address given below:
The National Simulation Resource, Director J. B. Bassingthwaighte, Department of Bioengineering, University of Washington, Seattle WA 98195-5061.
Model development and archiving support at https://www.imagwiki.nibib.nih.gov/physiome provided by the following grants: NIH U01HL122199 Analyzing the Cardiac Power Grid, 09/15/2015 - 05/31/2020, NIH/NIBIB BE08407 Software Integration, JSim and SBW 6/1/09-5/31/13; NIH/NHLBI T15 HL88516-01 Modeling for Heart, Lung and Blood: From Cell to Organ, 4/1/07-3/31/11; NSF BES-0506477 Adaptive Multi-Scale Model Simulation, 8/15/05-7/31/08; NIH/NHLBI R01 HL073598 Core 3: 3D Imaging and Computer Modeling of the Respiratory Tract, 9/1/04-8/31/09; as well as prior support from NIH/NCRR P41 RR01243 Simulation Resource in Circulatory Mass Transport and Exchange, 12/1/1980-11/30/01 and NIH/NIBIB R01 EB001973 JSim: A Simulation Analysis Platform, 3/1/02-2/28/07.

