Axially distributed 2-region capillary-tissue exchange operator for Albumin binding fatty acid at a particular binding site.
Description
One-dimensional convection-permeataion-diffusion- reaction model consisting of two concentric cylinders separated by a membrane. The central plasma region of volume Vp has flow Fp, first order consumption, Gp, and axial diffusion (disperion) Dp. Units are physiological per gram of tissue so that a single unit can model a homogeneously perfused organ. Radial diffusion is assumed instantaneous (short radial distances). Exchange into a second surrounding non-flowing region is passive with conductance, PS, the Permeability capillary Surface area product. The interstitial fluid region, isf, of volume Visf, like the capillary, is axially distributed, and the axial gradients are dissipated by concentration-independent axial diffusion or dispersion, D cm^2/s. Radial diffusion within this space is considered instantaneous, and consumption, Gisf, is first order. This model is used in multicapillary models as one of a set of units in parallel. Transcap extraction of F when there is only one, not multiple, independent binding sites on albumin. 1. Albumin flow is the driving event. Assume const flow. All sites move downstream together, same velocity. Piston flow. 2. Loss across endothelium has same params for all sites since the free F pool doesn't distinguish sites. 3. Assume equilibration BEFORE entry into capillary, i.e. the site occupancy is known from the I.C's 4. On-off equations for each site, exchanging with free F, will account for redistribution using the plasma concentration of F. (A shell of F in the immediate locale of Alb is NOT considered here.) Use the Independent Sites Model, not the Sequential Binding Model. 5. Capillary permeability is axially uniform, passive. 6. No return flux from tissue = Visf in region 2 is infinite. 7. Therefore need: 1 free alb, 1 free F, N binding sites with x-dependent occupancy. Minimum for 1-site: solute A, AF, and F, with only F permeating,i.e. Inputs for Cp, C1p, C1pF, FF simultaneous 8. Wish to determine the dependency of the transmembrane flux on concn. The expectation is that at High F, the off-rates of thelowest affinity sites will contribute most, and give the highest fluxes, but lowest F fluxes per unit free F, and lowest extraction fraction. Define extraction as the fraction of total F removed in single transcap passage. 9. Compare with 1983 data (Vusse etal.) re decrement in Extraction vs F with constant Atot/Ftot.
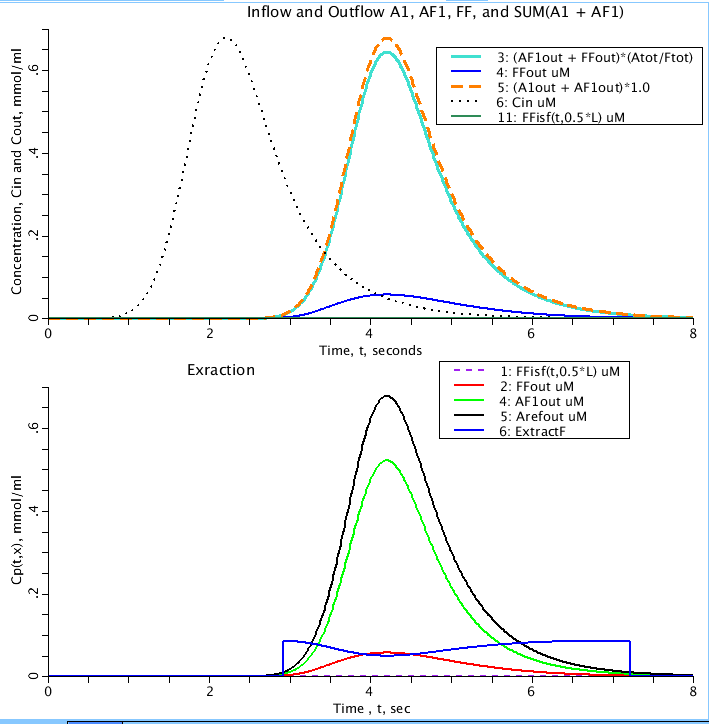
Figure: (Top) Inflow (Cin) and outflow of solutes through a capillary with exchange into the ISF of free fatty-acid (FF) and binding of FF to albumin (A). A1out is the conentration of albumin with site one free leaving the capillary. AF1out is the concentration of albumin with site one ocupied by FF leaving the capillary. FFisf is the concentration of FF in isf region at a point in the isf halfway down the capillary of length L. FFout is the concentration of FF leaving the capillary.Atot and Ftot are the total concentration of albumin and FF in teh system. Bottom Amount of free fatty-acid extracted (ExtractF) versus outflows as a function of time. Arefout is the reference albumin (Does not bind with FF) concentration leaving the capillary.
Equations
The equations for this model may be viewed by running the JSim model applet and clicking on the Source tab at the bottom left of JSim's Run Time graphical user interface. The equations are written in JSim's Mathematical Modeling Language (MML). See the Introduction to MML and the MML Reference Manual. Additional documentation for MML can be found by using the search option at the Physiome home page.
- Download JSim model MML code (text):
- Download translated SBML version of model (if available):
- No SBML translation currently available.
- Information on SBML conversion in JSim
We welcome comments and feedback for this model. Please use the button below to send comments:
Sangren WC and Sheppard CW. A mathematical derivation of the exchange of a labeled substance between a liquid flowing in a vessel and an external compartment. Bull Math Biophys 15: 387-394, 1953 (This gives an analytic solution for the two-region model.) Goresky CA, Ziegler WH, and Bach GG. Capillary exchange modeling: Barrier-limited and flow-limited distribution. Circ Res 27: 739-764, 1970. (This gives another derivation of the analytical form, and uses the model in both single and multicapillary models. Bassingthwaighte JB. A concurrent flow model for extraction during transcapillary passage. Circ Res 35: 483-503, 1974. (This gives numerical solutions, which are faster than the analytic solutions, and imbeds the model in an organ with tissue volumes conserved, and with arteries and veins.) Guller B, Yipintsoi T, Orvis AL, and Bassingthwaighte JB. Myocardial sodium extraction at varied coronary flows in the dog: Estimation of capillary permeability by residue and outflow detection. Circ Res 37: 359-378, 1975. (Application to sodium exchange in the heart.) Barta E, Sideman S, and Bassingthwaighte JB. Facilitated diffusion and membrane permeation of fatty acid in albumin solutions. Ann Biomed Eng 28: 331-345, 2000. Archived as www.imagwiki.nibib.nih.gov/physiome/jsim/ model 430 Equilibration calculation used in Barta et al (2000)
Please cite https://www.imagwiki.nibib.nih.gov/physiome in any publication for which this software is used and send one reprint to the address given below:
The National Simulation Resource, Director J. B. Bassingthwaighte, Department of Bioengineering, University of Washington, Seattle WA 98195-5061.
Model development and archiving support at https://www.imagwiki.nibib.nih.gov/physiome provided by the following grants: NIH U01HL122199 Analyzing the Cardiac Power Grid, 09/15/2015 - 05/31/2020, NIH/NIBIB BE08407 Software Integration, JSim and SBW 6/1/09-5/31/13; NIH/NHLBI T15 HL88516-01 Modeling for Heart, Lung and Blood: From Cell to Organ, 4/1/07-3/31/11; NSF BES-0506477 Adaptive Multi-Scale Model Simulation, 8/15/05-7/31/08; NIH/NHLBI R01 HL073598 Core 3: 3D Imaging and Computer Modeling of the Respiratory Tract, 9/1/04-8/31/09; as well as prior support from NIH/NCRR P41 RR01243 Simulation Resource in Circulatory Mass Transport and Exchange, 12/1/1980-11/30/01 and NIH/NIBIB R01 EB001973 JSim: A Simulation Analysis Platform, 3/1/02-2/28/07.

