This model simulates the electophysiological effect of inexcitable cardiac fibroblast when coupled with cardiac myocytes.
Description
This model simulates the role of cardiac fibroblasts on the action potential of cardiac myocytes when they are coupled together. The modulation of action potential can be explored by varying the number of fibroblasts coupled with a single myocyte and the intercellular resistance between myocytes and fibroblasts. The purpose of this model is to gain insights into the contribution of inexcitable cardiac fibroblasts to the electrophysiology of the myocardium. An electrophysiological fibroblast model developed by Sachse et al. and was coupled with the myocyte model of Pandit et al. The full model was created by Sachse et al. 2008.
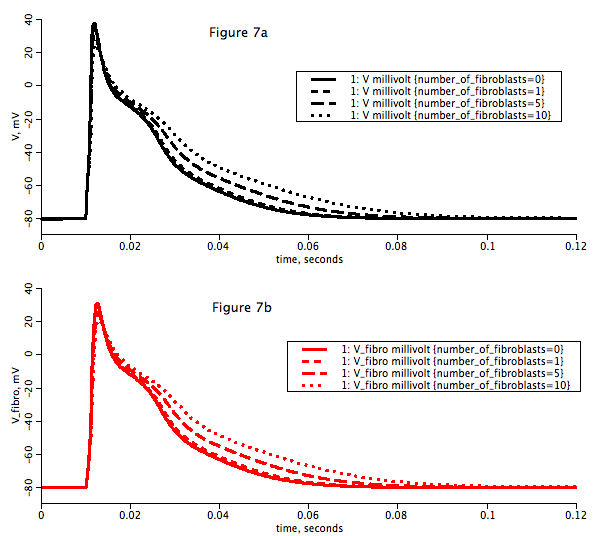
Figure: To reproduce figure 7, load model in JSim, select the 'Loops Run' from the 'All_Runs' pulldown menu and let the simulation run. To view the graph select the 'Figure_7ab' tab in the right panel of the JSim interaface.
Equations
The equations for this model may be viewed by running the JSim model applet and clicking on the Source tab at the bottom left of JSim's Run Time graphical user interface. The equations are written in JSim's Mathematical Modeling Language (MML). See the Introduction to MML and the MML Reference Manual. Additional documentation for MML can be found by using the search option at the Physiome home page.
- Download JSim model MML code (text):
- Download translated SBML version of model (if available):
We welcome comments and feedback for this model. Please use the button below to send comments:
Sachse FB, Moreno A, Abildskov JA.; Electrophysiological modeling of fibroblasts and their interaction with myocytes. Annals of Biomedical Engineering, 36(1): 41-56, 2008 Pandit SV, Clark RB, Giles WR, Demir SS. A mathematical model of action potential heterogeneity in adult left ventricular mycytes. Biophysical Journal 81:3029-3051, 2001.
Please cite https://www.imagwiki.nibib.nih.gov/physiome in any publication for which this software is used and send one reprint to the address given below:
The National Simulation Resource, Director J. B. Bassingthwaighte, Department of Bioengineering, University of Washington, Seattle WA 98195-5061.
Model development and archiving support at https://www.imagwiki.nibib.nih.gov/physiome provided by the following grants: NIH U01HL122199 Analyzing the Cardiac Power Grid, 09/15/2015 - 05/31/2020, NIH/NIBIB BE08407 Software Integration, JSim and SBW 6/1/09-5/31/13; NIH/NHLBI T15 HL88516-01 Modeling for Heart, Lung and Blood: From Cell to Organ, 4/1/07-3/31/11; NSF BES-0506477 Adaptive Multi-Scale Model Simulation, 8/15/05-7/31/08; NIH/NHLBI R01 HL073598 Core 3: 3D Imaging and Computer Modeling of the Respiratory Tract, 9/1/04-8/31/09; as well as prior support from NIH/NCRR P41 RR01243 Simulation Resource in Circulatory Mass Transport and Exchange, 12/1/1980-11/30/01 and NIH/NIBIB R01 EB001973 JSim: A Simulation Analysis Platform, 3/1/02-2/28/07.

