(Matlab) Model of open-loop (feed-forward) and feedback control of coronary blood flow during exercise, cardiac pacing, and pressure changes.
Description
(Abstract) A control system model was developed to analyze data on in vivo coronary blood flow regulation and to probe how different mechanisms work together to control coronary flow from rest to exercise, and under a variety of experimental conditions, including cardiac pacing and with changes in coronary arterial pressure (autoregulation). In the model coronary flow is determined by the combined action of a feedback pathway signal that is determined by the level of plasma ATP in coronary venous blood, an adrenergic open-loop (feed-forward) signal that increases with exercise, and a contribution of pressure-mediated myogenic control. The model was identified based on data from exercise experiments where myocardial oxygen extraction, coronary flow, cardiac interstitial norepinephrine concentration, and arterial and coronary venous plasma ATP concentrations were measured during control and during adrenergic and purinergic receptor blockade conditions. The identified model was used to quantify the relative contributions of open-loop and feedback pathways and to illustrate the degree of redundancy in the control of coronary flow. The results indicate that the adrenergic open-loop control component is responsible for most of the increase in coronary blood flow that occurs during high levels of exercise. However, the adenine nucleotide- mediated metabolic feedback control component is essential. The model was evaluated by predicting coronary flow in cardiac pacing and autoregulation experiments with reasonable fits to the data. The analysis shows that a model in which coronary venous plasma adenine nucleotides are a signal in local metabolic feedback control of coronary flow is consistent with the available data.
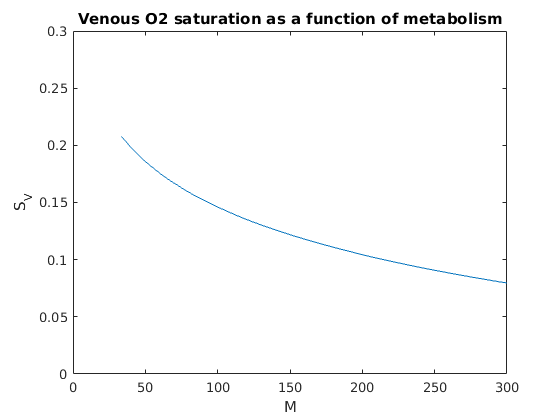
Figure 1: Relationship between metabolism, M (uL O2/(min*g)) and venous oxygen saturation (SV). From equation 6 of Pradhan et al. 2016: SV = SA - M/(F*Co*H), where SA is arterial oxygen saturation, F is coronary flow in ml/(min*g), Co is Oxygen content of blood when oxyhemoglobin is fully saturated and hematocrit is 100%, neglecting dissolved oxygen (uL O2/ml), H is hematocrit.
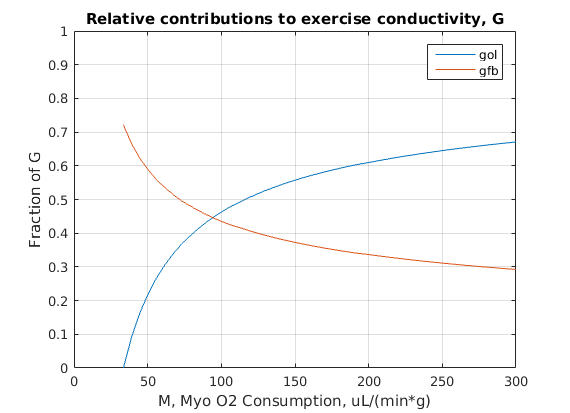
Figure 2: Effect of non basal conductance on coronary flow with changing metabolism, based on equation 11 of Pradhan et al. 2016, where Flow = G*P, P is mean arterial O2 pressure (mmHg) and G is total conductance (ml/(min*g*mmHg). G = g0 + gol + gfb, where g0 is basal conductance, gol is open-loop flow control pathway and gfb is feedback flow control pathway conductances.
Equations
The equations for this model may be viewed by opening up the Matlab model files downloaded from below.
Download Matlab model files
We welcome comments and feedback for this model. Please use the button below to send comments:
- (primary) Pradhan RK, Feigl EO, Gorman MW, Brengelmann GL, Beard DA Open-loop (feed-forward) and feedback control of coronary blood flow during exercise, cardiac pacing, and pressure changes, Am J Physiol Heart Circ Physiol 310: H1683–H1694, 2016. First published April 1, 2016; doi:10.1152/ajpheart.00663.2015
- Farias M 3rd, Gorman MW, Savage MV, Feigl EO. Plasma ATP during exercise: possible role in regulation of coronary blood flow. Am J Physiol Heart Circ Physiol 288: H1586–H1590, 2005
- Yada T, Richmond KN, Van Bibber R, Kroll K, Feigl EO. Role of adenosine in local metabolic coronary vasodilation. Am J Physiol Heart Circ Physiol 276: H1425–H1433, 1999
Please cite https://www.imagwiki.nibib.nih.gov/physiome in any publication for which this software is used and send one reprint to the address given below:
The National Simulation Resource, Director J. B. Bassingthwaighte, Department of Bioengineering, University of Washington, Seattle WA 98195-5061.
Model development and archiving support at https://www.imagwiki.nibib.nih.gov/physiome provided by the following grants: NIH U01HL122199 Analyzing the Cardiac Power Grid, 09/15/2015 - 05/31/2020, NIH/NIBIB BE08407 Software Integration, JSim and SBW 6/1/09-5/31/13; NIH/NHLBI T15 HL88516-01 Modeling for Heart, Lung and Blood: From Cell to Organ, 4/1/07-3/31/11; NSF BES-0506477 Adaptive Multi-Scale Model Simulation, 8/15/05-7/31/08; NIH/NHLBI R01 HL073598 Core 3: 3D Imaging and Computer Modeling of the Respiratory Tract, 9/1/04-8/31/09; as well as prior support from NIH/NCRR P41 RR01243 Simulation Resource in Circulatory Mass Transport and Exchange, 12/1/1980-11/30/01 and NIH/NIBIB R01 EB001973 JSim: A Simulation Analysis Platform, 3/1/02-2/28/07.

