Model the interactions of proteins cdc2 and cyclin which form a heterodimer (maturation promoting factor) that controls the major events of the cell cycle. Tyson 1991 model imported from Biomodels (www.ebi.ac.uk).
Description
(Abstract) The proteins cdc2 and cyclin form a heterodimer (maturation promoting factor) that controls the major events of the cell cycle. A mathematical model for the interactions of cdc2 and cyclin is constructed. Simulation and analysis of the model show that the control system can operate in three modes: as a steady state with high maturation promoting factor activity, as a spontaneous oscillator, or as an excitable switch. We associate the steady state with metaphase arrest in unfertilized eggs, the spontaneous oscillations with rapid division cycles in early embryos, and the excitable switch with growth-controlled division cycles typical of nonembryonic cells. Modeled as a set of six differential equations with the rate constant describing breakdown of the active cdc2-cyclin complex set to either constant value, or periodic (sine or exponential) with a period set to mass doubling time (cell division cycle).
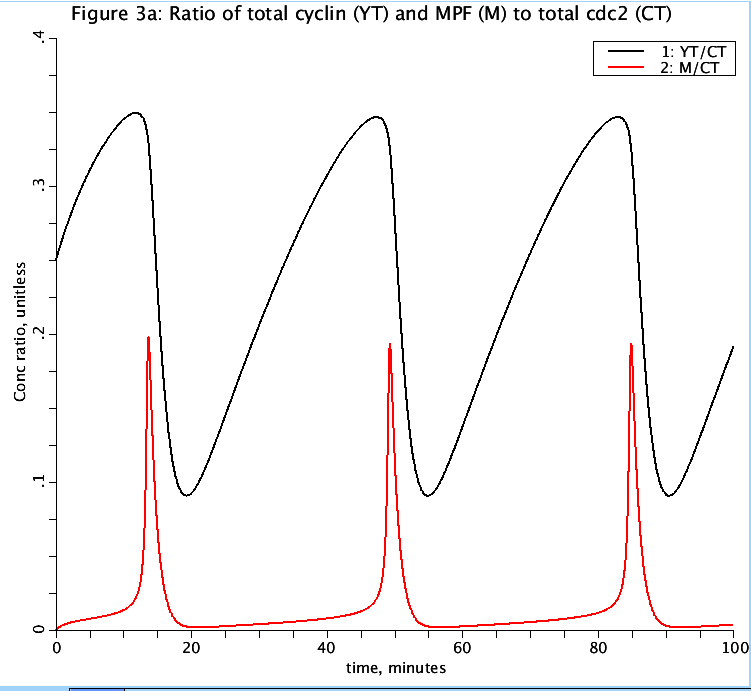
Figure: Dynamical behavior of the cdc2-cyclin model. Limit cycle oscillations for k4 = 180 min-1, k6 = 1 min^-1.
YT is total cyclin, CT is total cdc2, and M is active MPF.
Equations
The equations for this model may be viewed by running the JSim model applet and clicking on the Source tab at the bottom left of JSim's Run Time graphical user interface. The equations are written in JSim's Mathematical Modeling Language (MML). See the Introduction to MML and the MML Reference Manual. Additional documentation for MML can be found by using the search option at the Physiome home page.
- Download JSim model MML code (text):
- Download translated SBML version of model (if available):
We welcome comments and feedback for this model. Please use the button below to send comments:
Tyson JJ. Modeling the cell division cycle: cdc2 and cyclin interactions, 1991, www.ncbi.nlm.nih.gov/pubmed/1831270" www.ncbi.nlm.nih.gov/pubmed/20587024" ,Li C, Donizelli M, Rodriguez N, Dharuri H, Endler L, Chelliah V, Li L, He E, Henry A, Stefan MI, Snoep JL, Hucka M, Le Novre N, Laibe C (2010), BioModels Database: An enhanced, curated and annotated resource for published quantitative kinetic models. BMC Syst Biol., 4:92.
Please cite https://www.imagwiki.nibib.nih.gov/physiome in any publication for which this software is used and send one reprint to the address given below:
The National Simulation Resource, Director J. B. Bassingthwaighte, Department of Bioengineering, University of Washington, Seattle WA 98195-5061.
Model development and archiving support at https://www.imagwiki.nibib.nih.gov/physiome provided by the following grants: NIH U01HL122199 Analyzing the Cardiac Power Grid, 09/15/2015 - 05/31/2020, NIH/NIBIB BE08407 Software Integration, JSim and SBW 6/1/09-5/31/13; NIH/NHLBI T15 HL88516-01 Modeling for Heart, Lung and Blood: From Cell to Organ, 4/1/07-3/31/11; NSF BES-0506477 Adaptive Multi-Scale Model Simulation, 8/15/05-7/31/08; NIH/NHLBI R01 HL073598 Core 3: 3D Imaging and Computer Modeling of the Respiratory Tract, 9/1/04-8/31/09; as well as prior support from NIH/NCRR P41 RR01243 Simulation Resource in Circulatory Mass Transport and Exchange, 12/1/1980-11/30/01 and NIH/NIBIB R01 EB001973 JSim: A Simulation Analysis Platform, 3/1/02-2/28/07.

