A mathematical model of ventilation response to inhaled carbon monoxide, based on the work of James H. Stuhmiller and Louise M. Stuhmiller [2005, JAP, 98, 2033-2044] and developed by Raymond Yakura as a final project for BIOEN 589, University of Washington.
Description
A comprehensive mathematical model describing the respiration, circulation, oxygen metabolism,
and ventilatory control was assembled for the purpose of predicting acute ventilation changes
from exposure to carbon monoxide in both humans and animals. This Dynamic Physiological Model
is based on previously published work, reformulated, extended, and combined into a single model.
Model parameters are determined from literature values, fitted to experimental data, or
allometrically scaled between species. The model reaffirms the role of brain hypoxia on
hyperventilation during carbon monoxide exposures. Improvement in the estimation of total
ventilation, through a more complete knowledge of ventilation control mechanisms, and validated
by animal data, will increase the accuracy of inhalation toxicology estimates.
Introduction to the model:
With CO acute inhalation, hyperventilation first results and followed by
a subsequent ventilation depression. Hyperventilation is caused by hypoxia &
increased CO2 that activates the peripheral chemoreceptors. Then, ventilation
depression is caused by generation of lactic acid in the brain and by decreased
brain activity. A buildup of carboxyhemoglobin with reduction in oxygen
delivery to the brain leads to anaerobic glycolysis and build up of lactate.
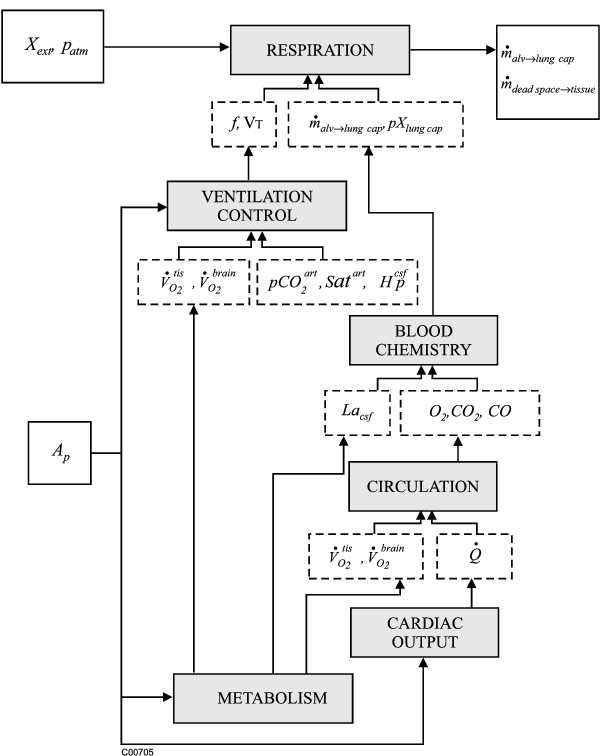
The model consists of 6 subsets:
(1) RESPIRATION SYSTEM which incompasses the total ventilation and
effects of dead space and humidification.
(2) VENTILATION which incompasses the chemoreceptor response,
brain activity response, and the combined ventilatory response
(3) CIRCULATORY SYSTEM which incompasses the mass balance equations
for O2, CO2 and CO,
(4) BLOOD CHEMISTRY which incompasses the hemoglobin saturation,
O2/CO partition, acid-base balance, and CO2 dissociation,
(5) CARDIAC OUTPUT which incompasses the blood flow to the brain
increases during hypoxia,
(6) METABOLIC SYSTEM which incompasses the oxygen metabolism, oxygen
transfer to the brain, lactic acid generation, and anaerobic limit,
Equations
The equations for this model may be viewed by running the JSim model applet and clicking on the Source tab at the bottom left of JSim's Run Time graphical user interface. The equations are written in JSim's Mathematical Modeling Language (MML). See the Introduction to MML and the MML Reference Manual. Additional documentation for MML can be found by using the search option at the Physiome home page.
- Download JSim model MML code (text):
- Download translated SBML version of model (if available):
We welcome comments and feedback for this model. Please use the button below to send comments:
Stuhmiller, J.H. and Stuhmiller, L.M.; A mathematical model of ventilation response to inhaled carbon monoxide, 2005, Journal of Applied Physiology, 98, 2033-2044. Ba ZF et al.; Alterations in tissue oxygen consumption and extraction after trauma and hemorrhagic shock. Critical Care Medicine 28(8), 2837-2842 (2000). Duffin J et al.; A mathematical model of the chemoreflex control of ventilation. Respir Physiol 120, 13-26 (2000). Gronlund et al.; Numerical Values of Classical Haldane Coefficient. JAP 57(3), 850-859 (1984).
Please cite https://www.imagwiki.nibib.nih.gov/physiome in any publication for which this software is used and send one reprint to the address given below:
The National Simulation Resource, Director J. B. Bassingthwaighte, Department of Bioengineering, University of Washington, Seattle WA 98195-5061.
Model development and archiving support at https://www.imagwiki.nibib.nih.gov/physiome provided by the following grants: NIH U01HL122199 Analyzing the Cardiac Power Grid, 09/15/2015 - 05/31/2020, NIH/NIBIB BE08407 Software Integration, JSim and SBW 6/1/09-5/31/13; NIH/NHLBI T15 HL88516-01 Modeling for Heart, Lung and Blood: From Cell to Organ, 4/1/07-3/31/11; NSF BES-0506477 Adaptive Multi-Scale Model Simulation, 8/15/05-7/31/08; NIH/NHLBI R01 HL073598 Core 3: 3D Imaging and Computer Modeling of the Respiratory Tract, 9/1/04-8/31/09; as well as prior support from NIH/NCRR P41 RR01243 Simulation Resource in Circulatory Mass Transport and Exchange, 12/1/1980-11/30/01 and NIH/NIBIB R01 EB001973 JSim: A Simulation Analysis Platform, 3/1/02-2/28/07.

