1. Model Overview
The chemistry in biological organisms is fundamentally different from that in test tubes and in chemical engineering largely due to the unique roles played by enzymes. With the presence of enzymes as specific catalysts, biochemical reactions are greatly accelerated. The unique role of enzymes also makes the kinetic of such reactions significantly different from that of conventional chemical kinetics. This was frist realized by L. Michaelis and M.L. Menten in 1913, when they developed a quantitative theory for the enzyme kinetics. As we shall see, the key difference between the enzyme kinetics and the conventional bimolecular reations is the large excess of substrate concentration with respect to the enzyme concentration.
We illustrate the uses of this kinetic model by beginning with an introduction and followed by several examples.
2. The Michaelis-Menten Kinetic Scheme and Equations
The core of the kinetic model MICMEN is a set of kinetic equations from which a set of ordinary differential equations can be derived. The differential equations then are solved numerically. The complete system of equations describe transient time courses for the concentrations of free substrate [S], bound substrate-enzyme complex [ES], and product [P] in well stirred compartments. These time courses are initiated by a set of initial concentrations not very different from a typical kinetic experiments. If one is interested in the steady-state of the reaction, one simply runs the kinetics and waits for the long-time behavior from the computations.
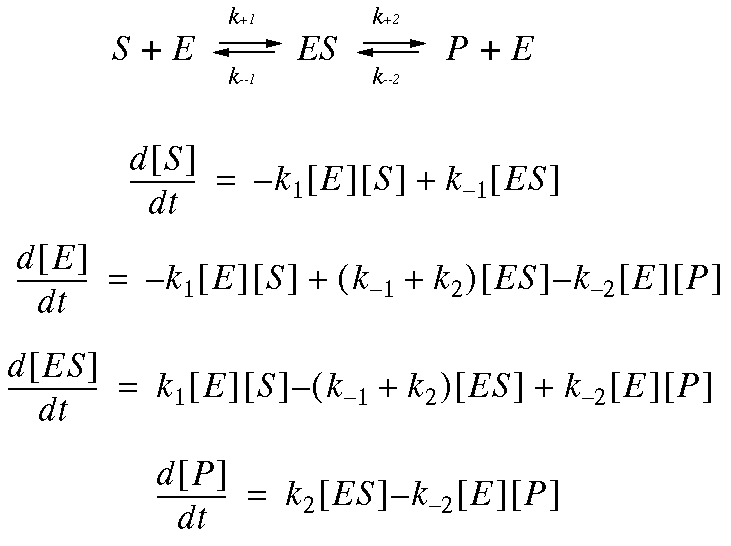
The above four equations are all based on the law of mass action which states that the rate of a reaction is proportional the the product of the concentrations of the reactants. The proportional constant is called a rate constant. In the above equation, k1 is called second-order while k2 is called first-order rate constants. It is easy to verify that ([E]+[ES]) is a constant in a reaction, so is ([S]+[ES]+[P]). They are the total enzyme concentration and total substrate concentration, respectively.
We now start the MICMEN program under XSIM. If one clicks on the "Parameters" button in the main manu and select the "Input" submanu, one will find a list of parameters for the MICMEN model. These parameters include 4 rate constants, and 4 initial concentrations at time zero. Giving a set of reasonable values, the MICMEN program will calculate the time evolution of the Michaelis-Menton (MM) reaction according to the kinetic scheme and equations.
3. A Comparison Between MM Kinetics and Conventional Bimolecular Kinetics
Usually a reaction starts with ES0 = P0 = 0. If the enzyme concentration are significantly greater than that of substrate, (e.g., E0=100, S0=1), then the reaction is essentially an exponentially decreasing of [S] and increasing of [ES]. Run the example ex1.par. After the main XSIM window is sent to your screen, you can click on the "Run" button to start the model similation. While the model is running, click on the "Results" button in the main manu and select the "Plot Area 1", you will see how the concentrations [S] and [ES] are progressing. If you change the Y-axis from linear to log, you will see the [S] is decreasing in a perfect exponential fashion.
If now you change S0 to 100 and E0 to 1, and run the simulation again, you will see a completely different picture. The reaction is much slower and within the total time of 0.05, there is essentially no changes in the [S]. To see how [ES] is changing, you change the ES in the "Y Parameters" window under the "Plot_Area 1" to ES*100. What you see is that while [S] is essentially unchanged, [ES] is increasing exponentially.
Now for a more complete picture of the reaction, click on the "Parameters" bottom in the main manu and select the "time" submanu. Change the "stop" to 50, and the "Incr" to 0.1, and run the simulation again.
4. Steady-State Kinetics and Its Validity
What one sees is that after the brief transient, the [ES] is kept at a constant level while [S] is decreasing linearly. If you also plot [P], it is increasing linearly. This situation is known as pseudo-steady-state in which the substrate is converted into production with a constant rate, while [ES] is kept at a constant "steady-state" level, approximately = E0. This phenomenon is called saturation: the enzyme is saturated by the substrate and becomes a bottleneck for the entire reaction. This is the essence of MM kinetics which is fundamentally different from a conventional chemical kinetics in which usually there are equal amount of reactants.
If we change the "stop" to 500, and the "Incr" to 1, and run the simulation again, we see that eventially [S] and [P] reach an equilibrium with 1:1 ratio. This is expected since all the k's are 1. We now have a complete picture of the MM reaction which has three time regimes: a transient, a pseudo-steady-state, and an equilibrium.
5. Rapid Equilibrium and Steady-State Kinetics
For different sets of rate constants, the MM reaction can behave differently. The second example ex2.par illustrates the situation known as rapid equilibrium. In this case, the k-1 are significantly greater than k2. Hence the reaction within the first step (binding) is esseitally at it equilibrium all the time, with minor turnover from ES to P. One can see that the ratio [E][S]/[ES] are at its equilibrium value (k-1/k+1) at all time during the reaction. Both E and ES are decreasing with tha rate k2[ES]. In the current example, we have set k-2=0, hence the reaction will stop only when all the substrate is converted into the product.
6. References
Fersht, A. Enzyme Structure and Mechanism.W. H. Freeman, San Francisco, (1977).
Hammes, G.G. Enzyme Catalysis and Regulation. Academic Press, New York, (1982).
Murray, J.D. Mathematical Biology.Springer-Verlag, New York, (1993).
Segel, I.H. Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-State Enzyme Systems. John-Wiley & Sons, New York, (1975).

