TABLE OF CONTENTS
Model demonstrations with parameter sets
1. IMPORTANT !!! To run a demonstration, enter "xhost +nsr.bioeng.washington.edu" in one of your windows to allow the demonstration program to open windows on your machine !!!
2. Parameters
SIMVOL is parameterized as follows.
Table 1. SIMVOL Parameters
Access to all input variables is via clicking the "INPUT" button on the main model page or by selecting "Inputs" under the "Parameters" menu.
3. Output Variables
Output variables are given by:
Table 2. SIMVOL Output Variables
|
Amount of material recirculation back into the system in the next time step |
To plot results, select "Plot Area 1" from the Results menu. Place the cursor in an unused box under Y-parameters and press the right mouse button to bring up a list of plottable parameters.
4. Diagram
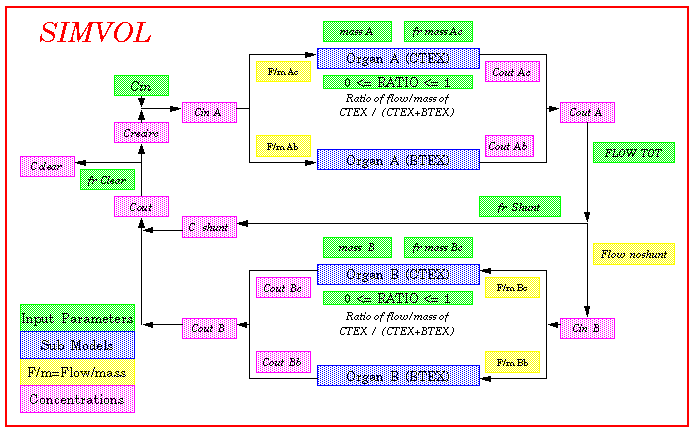
5. Model demonstrations with parameter sets
The user should inspect the input parameters and the output curves. Output curves can be found under the Results button under Plot Area 1 and Plot Area 2.
The following parameter sets are can be demonstrated by clicking on them:
5.1. Figure 9.2 : Figure09.02.par
Run the model. Displacement of fluid from a well stirred chamber. Flow is 600 ml/min (10 ml/sec). Volume is 100 ml. Amount of material injected is 1 mmole. Numeric and analytic solutions are plotted.
5.2. Figure 9.3: Figure09.03.par
Run the model. Dilution into a pair of well stirred volumes. Time course of concentrations with pulse input when each of V1 and V2 are instaneously mixed. (Flow=10 ml/sec, V1=100ml, V2=200ml)
5.3. Figure 9.5: Figure09.05.par
Run the model. See output under Results: Plot Area 1. The response of the system when the solute is constrained to just the plasma space, compared to the solute entering the ISF space, with concentrations measured at the outflows of this two-organ "whole-body" model.
5.4. Figure 9.6: Figure09.06.par
Run the model. Tracer concentration-time courses under Results: Plot Area 1 for 4-organ, 3-tissue-region model analogous to that in Figure 9.5 (zero clearance). Each "organ" was composed of regions for plasma, ISF, and cells. An extracellular flow marker enters the former two regions only; tracer water enters all three regions.
5.5. Figure 9.7: Figure09.07.par
Run the model. Plasma concentratime time curves for plasma, extracellular flow, and water markers where there is tracer lost from the body and the clearance is 30%.
5.6. Figure 9.8: Figure09.08.par
Semilog plots of plasma concentrations of markers for plasma, ECF and water spaces when clearance is exactly 30% of the flow or "cardiac output" in the model used in Figure 9.6. All curves have a monoexponential tail.
5.7. Figure 9.9: Figure09.09
Semilog plot of plasma concentration time curves for a strictly intravascular marker which is gradually cleared from the circulation. The tails of the curves are fitted by a single exponential. The loss rate or clearance was set at 0, 5, 10, 15, and 20% of the whole body recirculation. Monoexponential curves fitting the "data" from 100 to 200 seconds extrapolate back to an apparent common intersection at negative time. The intercept at t=0 is lower with clearance > 0, and using its value, C(t=0) , gives overestimates of Vpl .
Copyright © 2000, National Simulation Resource, University of Washington.
Contact garyr@nsr.bioeng.washington.edu with comments, questions, or critiques.

