Michaelis-Menten transporter models.
These are a takeoff on Michaelis-Menten characterization of enzyme kinetics. As such, the models assume instantaneous binding of solute to transporter, and assume transporter concentrations are negligible relative to the solute concentration, There is no accounting for the mass of solute bound to transporter. These are "saturable" transporters, meaning that at high solute concentrations all of the transporter molecules have their active sites filled by solute molecules and the rate of transport reaches a maximum. Thus transport is "concentration-dependent".
Like enzymatic reactions, the flux is bidirectional. Unlike enzymes the "substrate" and "product" are identical. Unlike enzymes there is sidedness, and the substrate can be bound on either side of the membrane. Since in actuality a transporter molecule exposes its active site to only one side at a time, and the concentration of transporter-substrate complex on each particular side provides the driving force for the particular flux there arises conflicts among the assumptions.
Cautionary Note:
The enzyme formulation of Michaelis-Menten kinetics has the rate constant
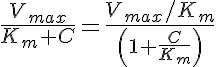
However, for consistency in units in the transporter formulation, we define the MM transporter rate constant as
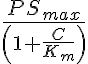
where PSmax equals Vmax divided by the Michaelis constant Km, and PSmax has the same units as the flow, exchange, and consumption rates. In some of these models PSmax will be an output variable and in some of these models it will be an input variable.
Michaelis-Menten Models:
- One sided 2 compartment.
- One sided 2 compartment with flow.
- One sided 2 region with flow.
- Two sided 2 compartment.
- Two sided 2 compartment with flow.
- Two sided 2 region with flow.
- Two sided 3 region, 2 transporters with flow.
References
Bassingthwaighte JB. A concurrent flow model for extraction during transcapillary passage. Circ Res 35: 483-503, 1974. (This gives numerical solutions, which are faster than the analytic solutions, and imbeds the model in an organ with tissue volums conserved, and with arteries and veins. The original Lagrangian sliding fluid element model with diffusion.)
Klingenberg M. Membrane protein oligomeric structure and transport function. Nature 290: 449-454, 1981.
Stein WD. The Movement of Molecules across Cell Membranes. New York: Academic Press, 1967.
Stein WD. Transport and Diffusion across Cell Membranes. Orlando, Florida: Academic Press Inc., 1986.
Wilbrandt W and Rosenberg T. The concept of carrier transport and its corollaries in pharmacology. Pharmacol Rev 13: 109-183, 1961.
Schwartz LM, Bukowski TR, Ploger JD, and Bassingthwaighte JB. Endothelial adenosin transporter characterization in perfused guinea pig hearts. Am J Physiol Heart Circ Physiol 279: H1502-H1511, 2000.
Dawson CA, Linehan JH, Rickaby DA, and Roerig DL. Influence of plasma protein on the inhibitory effects of indocyanine green and bromcresol green on pulmonary prostaglandin E1 extraction. Br J Pharmac 81: 449-455, 1984.
Crone C. Facilitated transfer of glucose from blood into brain tissue. J Physiol 181: 103-113, 1965.
Key Terms
Michaelis-Menten, bidirectional, saturable transporter, instantaneous binding.
JSim Tutorial
Click here to go to a JSim tutorial webpage, with an introduction to the JSim GUI, detailed usage instructions, and an accompaying video.
Copyright (C) 1999-2009 University of Washington. From the National Simulation Resource, Director J. B. Bassingthwaighte, Department of Bioengineering, University of Washington, Seattle WA 98195-5061. Academic use is unrestricted. Software may be copied so long as this copyright notice is included. This software was developed with support from NIH grant HL073598. Please cite this grant in any publication for which this software is used and send one reprint to the address given above.
Model development and archiving support at https://www.imagwiki.nibib.nih.gov/physiome provided by the following grants: NIH U01HL122199 Analyzing the Cardiac Power Grid, 09/15/2015 - 05/31/2020, NIH/NIBIB BE08407 Software Integration, JSim and SBW 6/1/09-5/31/13; NIH/NHLBI T15 HL88516-01 Modeling for Heart, Lung and Blood: From Cell to Organ, 4/1/07-3/31/11; NSF BES-0506477 Adaptive Multi-Scale Model Simulation, 8/15/05-7/31/08; NIH/NHLBI R01 HL073598 Core 3: 3D Imaging and Computer Modeling of the Respiratory Tract, 9/1/04-8/31/09; as well as prior support from NIH/NCRR P41 RR01243 Simulation Resource in Circulatory Mass Transport and Exchange, 12/1/1980-11/30/01 and NIH/NIBIB R01 EB001973 JSim: A Simulation Analysis Platform, 3/1/02-2/28/07.

