A two compartment two-sided Michaelis-Menten transporter with two solutes.
Detailed Description
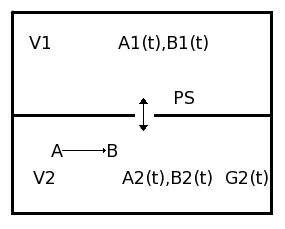
A facilitated transporter kinetic model assuming instantaneous solute binding to transporter of Michaelis-Menten type, single site available from both sides of the membrane. This model contains two solutes in two compartments, giving concentrations A1, A2, B1, and B2. Permeability-Surface Area product (PS) is determined by the concentration of A1 and B2. PSmax is max PS at 0% saturation. There is Gulosity in V2. In V2, A is consumed, forming B. B is also consumed.
Relevant Equations
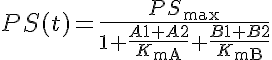
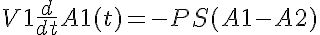
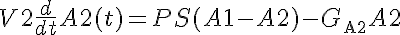
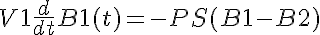
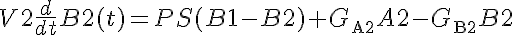
KmA and KmB are the equilibrium dissociation constants for each solute on both sides.
We welcome comments and feedback for this model. Please use the button below to send comments:
Klingenberg M. Membrane protein oligomeric structure and transport function. Nature 290: 449-454, 1981.
Stein WD. The Movement of Molecules across Cell Membranes. New York: Academic Press, 1967.
Stein WD. Transport and Diffusion across Cell Membranes. Orlando, Florida: Academic Press Inc., 1986.
Wilbrandt W and Rosenberg T. The concept of carrier transport and its corollaries in pharmacology. Pharmacol Rev 13: 109-183, 1961.
Schwartz LM, Bukowski TR, Ploger JD, and Bassingthwaighte JB. Endothelial adenosin transporter characterization in perfused guinea pig hearts. Am J Physiol Heart Circ Physiol 279: H1502-H1511, 2000.
Foster DM and Jacquez JA. An analysis of the adequacy of the asymmetric carrier model for sugar transport. Biochim Biophys Acta 436: 210-221, 1976.
Copyright (C) 1999-2010 University of Washington. From the National Simulation Resource, Director J. B. Bassingthwaighte, Department of Bioengineering, University of Washington, Seattle WA 98195-5061. Academic use is unrestricted. Software may be copied so long as this copyright notice is included. This software was developed with support from NIH grant HL073598. Please cite this grant in any publication for which this software is used and send one reprint to the address given above.
Model development and archiving support at https://www.imagwiki.nibib.nih.gov/physiome provided by the following grants: NIH U01HL122199 Analyzing the Cardiac Power Grid, 09/15/2015 - 05/31/2020, NIH/NIBIB BE08407 Software Integration, JSim and SBW 6/1/09-5/31/13; NIH/NHLBI T15 HL88516-01 Modeling for Heart, Lung and Blood: From Cell to Organ, 4/1/07-3/31/11; NSF BES-0506477 Adaptive Multi-Scale Model Simulation, 8/15/05-7/31/08; NIH/NHLBI R01 HL073598 Core 3: 3D Imaging and Computer Modeling of the Respiratory Tract, 9/1/04-8/31/09; as well as prior support from NIH/NCRR P41 RR01243 Simulation Resource in Circulatory Mass Transport and Exchange, 12/1/1980-11/30/01 and NIH/NIBIB R01 EB001973 JSim: A Simulation Analysis Platform, 3/1/02-2/28/07.

