Four region, distributed model for the O2-CO2 transport, exchange and metabolism. From Dash 2006 paper. Annotated for use with MPC. Compute dissolved O2 and CO2 from total O2 and CO2 (TO2 and TCO2) through numerical inversion method that uses SHbO2CO2_EJAP2016 routine and Christmas 2017 O2/CO2 solubility algorithm. Incorporates Calculations for temperature changes based on consumption of O2 in parenchymal region.
Description
Four region model for the O2-CO2 transport, exchange and metabolism. Based on Dash et al 2006 and 2010 papers. Computes dissolved O2 and CO2 from total O2 and CO2 (TO2 and TCO2) through numerical inversion method using Dash et al. 2016 routine. O2 and CO2 gas solubility change with time and position based on Christmas et al. 2017 O2/CO2 solubility algorithm that incorporates temperature and density (or water space) into its solubility calculations. Incorporates calculations for temperature changes (time and position dependent) based on consumption of O2 generating heat in parenchymal cell (PC) region. Model incorporated into larger recirculating model (Model #405, RecircO2CO2BTEX_3CompLung).
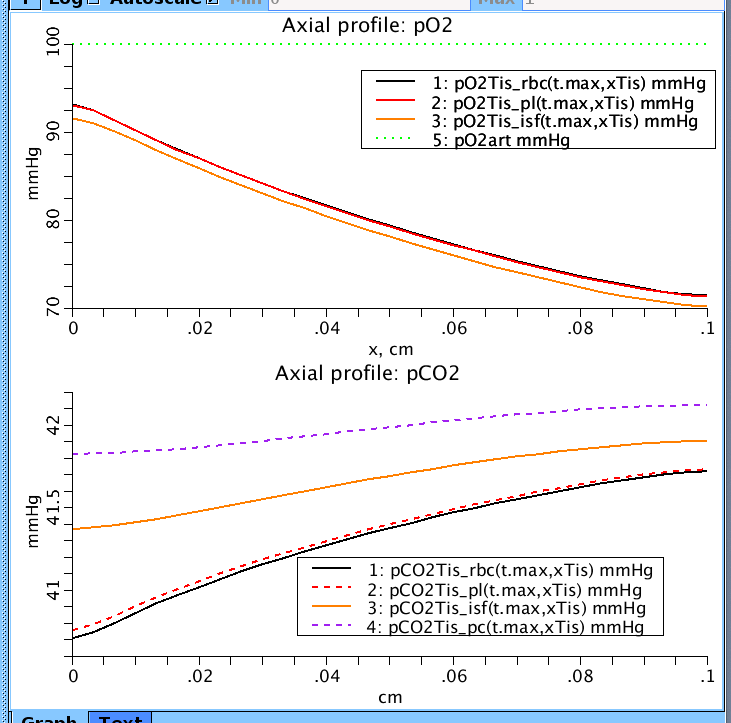
Figure 1: Axial distribution of pO2 and pCO2 along tissue axis at t.max=100 s. rbc = red blood cells, pl = plasma region, isf = interstitial region, and pc = parenchymal cells (musscle cells). pO2 and pCO2 going into tissue: 100 mmHg and 40 mmHg respectively.
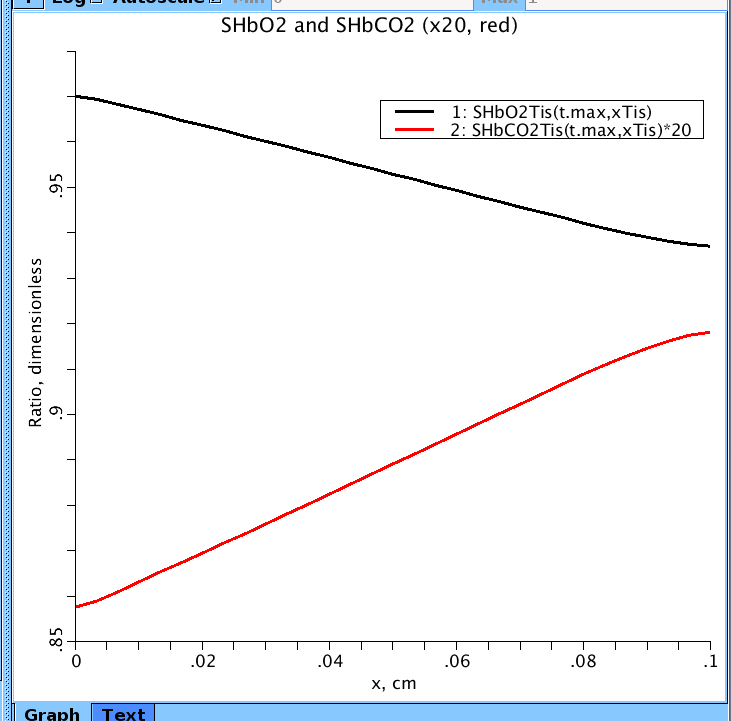
Figure 2: Axial distribution of hemoglobin saturation SHbO2(Blk) and SHbCO2(x20) along tissue axis at t.max=100 s. rbc = red blood cells, pl = plasma region, isf = interstitial region, and pc = parenchymal cells (musscle cells). pO2 and pCO2 going into tissue: 100 mmHg and 40 mmHg respectively.
Equations
The equations for this model may be viewed by running the JSim model applet and clicking on the Source tab at the bottom left of JSim's Run Time graphical user interface. The equations are written in JSim's Mathematical Modeling Language (MML). See the Introduction to MML and the MML Reference Manual. Additional documentation for MML can be found by using the search option at the Physiome home page.
- Download JSim model MML code (text):
- Download translated SBML version of model (if available):
- No SBML translation currently available.
- Information on SBML conversion in JSim
We welcome comments and feedback for this model. Please use the button below to send comments:
Yipintsoi T, Scanlon PD, and Bassingthwaighte JB. Density and water content of dog ventricular
myocardium. Proc Soc Exp Biol Med 141: 1032-1035, 1972.
Bassingthwaighte JB, Yipintsoi T, and Harvey RB. Microvasculature of the dog left ventricular
myocardium. Microvasc Res 7: 229-249, 1974.
Gonzalez F and Bassingthwaighte JB. Heterogeneities in regional volumes of distribution and
flows in the rabbit heart. Am J Physiol Heart Circ Physiol 258: H1012-H1024, 1990.
Vinnakota K and Bassingthwaighte JB. Myocardial density and composition: A basis for
calculating intracellular metabolite concentrations.
Am J Physiol Heart Circ Physiol 286: H1742-H1749, 2004.
Bassingthwaighte JB and Vinnakota KC. The computational integrated myocyte.
A view into the virtual heart. In: Modeling in Cardiovascular Systems.
Ann. New York Acad. Sci. 1015:, edited by Sideman S. and Beyar R. 2004, pp 391-404.
Brozek J, Grande F, Anderston JT, and Keys A. Densitometric analysis of body composition: revision of
some quantitative assumptions. Ann NY Acad Sci 110: 113–140, 1963.
Effros RM and Chinard FP. The in vivo pH of the extravascular space
of the lung. J Clin Invest 48: 1983–1996, 1969. (for water content of RBC)
Haurowitz, F. Chemistry and Biology of Proteins. New York: Academic Press; 1950. (for rho protein)
Allen TH, Krzywicki HJ, Roberts JE. J. Appl. Physiol 1959;14:1005. [PubMed: 13792786]
Allen TH, Welch BE, Trujillo TT, Roberts JE. J. Appl. Physiol 1959;14:1009. [PubMed: 13792787]
Please cite https://www.imagwiki.nibib.nih.gov/physiome in any publication for which this software is used and send one reprint to the address given below:
The National Simulation Resource, Director J. B. Bassingthwaighte, Department of Bioengineering, University of Washington, Seattle WA 98195-5061.
Model development and archiving support at https://www.imagwiki.nibib.nih.gov/physiome provided by the following grants: NIH U01HL122199 Analyzing the Cardiac Power Grid, 09/15/2015 - 05/31/2020, NIH/NIBIB BE08407 Software Integration, JSim and SBW 6/1/09-5/31/13; NIH/NHLBI T15 HL88516-01 Modeling for Heart, Lung and Blood: From Cell to Organ, 4/1/07-3/31/11; NSF BES-0506477 Adaptive Multi-Scale Model Simulation, 8/15/05-7/31/08; NIH/NHLBI R01 HL073598 Core 3: 3D Imaging and Computer Modeling of the Respiratory Tract, 9/1/04-8/31/09; as well as prior support from NIH/NCRR P41 RR01243 Simulation Resource in Circulatory Mass Transport and Exchange, 12/1/1980-11/30/01 and NIH/NIBIB R01 EB001973 JSim: A Simulation Analysis Platform, 3/1/02-2/28/07.

