A mathematical model of metabolic insulin signaling pathways based on the work of Ahmad Sedaghat, Arthur Sherman and Michael Quon [2002, AM J Physiol Endocrinol Metab, 283,E1084-E1101].
Description
This is a mathematical model that explicitly represents many of the known
signaling components mediating translocation of the insulin-responsive glucose
transporter GLUT4 and gives insight into the complexities of metabolic insulin
signaling pathways. A novel mechanistic model of postreceptor events including
phosphorylation of insulin receptor substrate-1,activation of phosphatidylinositol
3-kinase, and subsequent activation of downstream kinases Akt and protein kinase C
is coupled with previously validated subsystem models of insulin receptor binding,
receptor recycling, and GLUT4 translocation. A system of differential equations is
defined by the structure of the model. Rate constants and model parameters are
constrained by published experimental data. Model simulations of insulin
dose-response experiments agree with published experimental data and also generate
expected qualitative behaviors such as sequential signal amplification and increased
sensitivity of downstream components.
This JSim version was developed by Maris Lemba [2003] as a project while at the University of Washington.
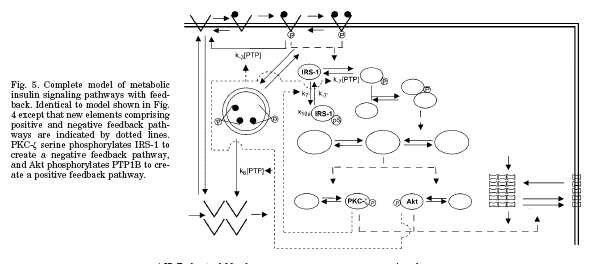
Figure 1: Diagram of model with feedback. From Sedaghat et al. 2002 paper.
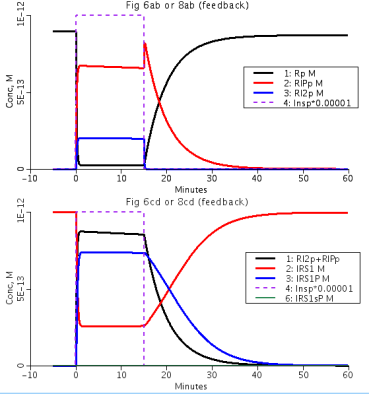
Figure 2: No feedback. Figure 6 from Sedaghat et al. 2002 paper. Time course for (Top) unbounded receptors (Rp), once and twice-bound phosphorylated surface receptors (RIPp and RI2p),(bottom) total phosphorylated surface receptors (RI2p+RIPp), and unphosphorylated (IRS1) and tyrosine-phosphorylated IRS-1 after a step input of insulin fo 15 min. Purple dashed line is insulin input pulse.
Equations
The equations for this model may be viewed by running the JSim model applet and clicking on the Source tab at the bottom left of JSim's Run Time graphical user interface. The equations are written in JSim's Mathematical Modeling Language (MML). See the Introduction to MML and the MML Reference Manual. Additional documentation for MML can be found by using the search option at the Physiome home page.
- Download JSim model MML code (text):
- Download translated SBML version of model (if available):
We welcome comments and feedback for this model. Please use the button below to send comments:
Ahmad Sedaghat, Arthur Sherman and Michael Quon, A mathematical model of metabolic
insulin signaling pathways, 2002, AM J Physiol Endocrinol Metab, 283,E1084-E1101.
Please cite https://www.imagwiki.nibib.nih.gov/physiome in any publication for which this software is used and send one reprint to the address given below:
The National Simulation Resource, Director J. B. Bassingthwaighte, Department of Bioengineering, University of Washington, Seattle WA 98195-5061.
Model development and archiving support at https://www.imagwiki.nibib.nih.gov/physiome provided by the following grants: NIH U01HL122199 Analyzing the Cardiac Power Grid, 09/15/2015 - 05/31/2020, NIH/NIBIB BE08407 Software Integration, JSim and SBW 6/1/09-5/31/13; NIH/NHLBI T15 HL88516-01 Modeling for Heart, Lung and Blood: From Cell to Organ, 4/1/07-3/31/11; NSF BES-0506477 Adaptive Multi-Scale Model Simulation, 8/15/05-7/31/08; NIH/NHLBI R01 HL073598 Core 3: 3D Imaging and Computer Modeling of the Respiratory Tract, 9/1/04-8/31/09; as well as prior support from NIH/NCRR P41 RR01243 Simulation Resource in Circulatory Mass Transport and Exchange, 12/1/1980-11/30/01 and NIH/NIBIB R01 EB001973 JSim: A Simulation Analysis Platform, 3/1/02-2/28/07.

