Simulation of oxyhemoglobin (HbO2) and carbaminohemoglobin (HbCO2) dissociation curves and computation of total O2 and CO2 contents in RBCs, Modified from Dash's original 2016 Matlab version. Annotated for use with MPC.
Description
-----------------------------------------------------------------------------------------
Simulation of oxyhemoglobin (HbO2) and carbaminohemoglobin (HbCO2) dissociation
curves and computation of total O2 and CO2 contents in whole blood, revised from
the original model of Dash and Bassingthwaighte, ABME 38(4):1683-1701, 2010. The
revision makes the model further simplified, as it bipasses the computations of
the indices n1, n2, n3 and n4, which are complex expressions. Rather the revision
necessitates the computations of K4p in terms of P50. Also the calculations of P50
in terms of pH is enhanced based on a 3rd degree polynomial interpolation.
-----------------------------------------------------------------------------------------
Original Matlab version developed by: Ranjan Dash, PhD (Last modified: 2/29/2016 from 3/15/2015 version)
Department of Physiology and Biotechnology and Bioengineering Center
Medical College of Wisconsin, Milwaukee, WI-53226
-----------------------------------------------------------------------------------------
BIOCHEMICAL REACTIONS FOR DERIVATION OF THE SHBO2 AND SHBCO2 EQUATIONS----------------
The equations for O2 and CO2 saturations of hemoglobin (SHbO2 and SHbCO2) are
derived by considering the various kinetic reactions involving the binding of
O2 and CO2 with hemoglobin in RBCs:
kf1p K1dp
1. CO2+H2O <--> H2CO3 <--> HCO3- + H+; K1_rbc=(kf1p/kb1p)*K1dp
kb1p K1 = 7.43e-7 M; K1dp = 5.5e-4 M
kf2p K2dp
2. CO2+HbNH2 <--> HbNHCOOH <--> HbNHCOO- + H+; K2=(kf2p/kb2p)*K2dp
kb2p K2 = 21.5e-6; K2dp = 1.0e-6 M
kf3p K3dp
3. CO2+O2HbNH2 <--> O2HbNHCOOH <--> O2HbNHCOO- + H+; K3=(kf3p/kb3p)*K3dp
kb3p K3 = 11.3e-6; K3dp = 1.0e-6 M
kf4p
4. O2+HbNH2 <--> O2HbNH2; K4p=func([O2];[H+];[CO2];[DPG];T)
kb4p
K5dp
5. HbNH3+ <--> HbNH2 + H+; K5dp = 2.4e-8 M
K6dp
6. O2HbNH3+ <--> O2HbNH2 + H+; K6dp = 1.2e-8 M
Download Dash et al. 2016 paper.
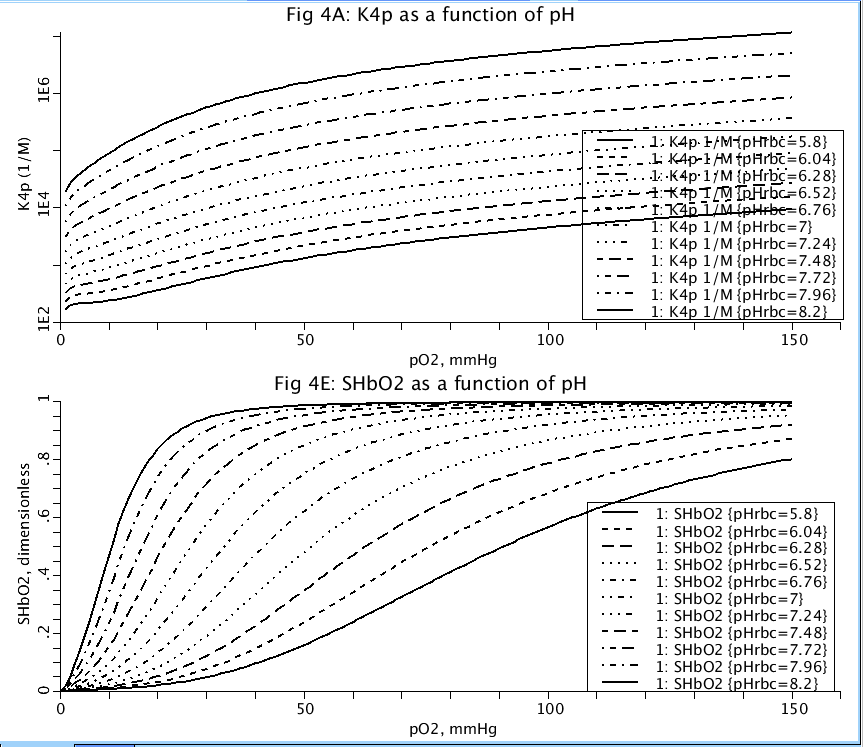
Figure: Top (Fig4A), Equilibrium constant for uptake of O2 by hemoglobin (K4p) as a function of pH and partial pressure of Oxygen in the red blood cells. Bottom (Fig4E), Saturation of hemoglobin by oxygen (SHbO2) as a function of pH and partial pressure of Oxygen in the red blood cells.
Equations
The equations for this model may be viewed by running the JSim model applet and clicking on the Source tab at the bottom left of JSim's Run Time graphical user interface. The equations are written in JSim's Mathematical Modeling Language (MML). See the Introduction to MML and the MML Reference Manual. Additional documentation for MML can be found by using the search option at the Physiome home page.
- Download JSim model MML code (text):
- Download translated SBML version of model (if available):
We welcome comments and feedback for this model. Please use the button below to send comments:
(primary) Dash, R.K., Korman, B. & Bassingthwaighte, J.B. Simple accurate mathematical models of blood HbO2 and HbCO2 dissociation curves at varied physiological conditions: evaluation and comparison with other models. Eur J Appl Physiol (2016) 116: 97. doi:10.1007/s00421-015-3228-3 Dash RK and Bassingthwaighte JB. Erratum to: Blood HbO2 and HbCO2 dissociation curves at varied O2, CO2, pH, 2,3-DPG and Temperature Levels. Ann Biomed Eng 38(4): 1683-1701, DOI: 10.1007/s10439-010-9948-y PMC2862600, 2010 Dash RK, Bassingthwaighte JB (2006) Simultaneous blood-tissue exchange of oxygen, carbon dioxide, bicarbonate, and hydrogen ion. Ann Biomed Eng 34:1129–1148
Please cite https://www.imagwiki.nibib.nih.gov/physiome in any publication for which this software is used and send one reprint to the address given below:
The National Simulation Resource, Director J. B. Bassingthwaighte, Department of Bioengineering, University of Washington, Seattle WA 98195-5061.
Model development and archiving support at https://www.imagwiki.nibib.nih.gov/physiome provided by the following grants: NIH U01HL122199 Analyzing the Cardiac Power Grid, 09/15/2015 - 05/31/2020, NIH/NIBIB BE08407 Software Integration, JSim and SBW 6/1/09-5/31/13; NIH/NHLBI T15 HL88516-01 Modeling for Heart, Lung and Blood: From Cell to Organ, 4/1/07-3/31/11; NSF BES-0506477 Adaptive Multi-Scale Model Simulation, 8/15/05-7/31/08; NIH/NHLBI R01 HL073598 Core 3: 3D Imaging and Computer Modeling of the Respiratory Tract, 9/1/04-8/31/09; as well as prior support from NIH/NCRR P41 RR01243 Simulation Resource in Circulatory Mass Transport and Exchange, 12/1/1980-11/30/01 and NIH/NIBIB R01 EB001973 JSim: A Simulation Analysis Platform, 3/1/02-2/28/07.

