Models the adaptation of ATP production by the mitochondria to ATP hydrolysis. This model contains only diffusion, mitochondrial outer membrane (MOM) permeation, and two isoforms of creatine kinase (CK), in cytosol and mitochondrial intermembrane space (IMS), respectively.
Description
ADENINE NUCLEOTIDE-CREATINE PHOSPHATE MODULE IN METABOLIC SYSTEM EXPLAINS FAST PHASE OF DYNAMIC REGULATION OF OXIDATIVE PHOSPHORYLATION. Computational models of a large metabolic system can be assembled from modules that represent a biological function emerging from interaction of a small subset of molecules. A “skeleton model” is tested here for a module that regulates the first phase of dynamic adaptation of oxidative phosphorylation (OxPhos) to demand in heart muscle cells. The model contains only diffusion, mitochondrial outer membrane (MOM) permeation, and two isoforms of creatine kinase (CK), in cytosol and mitochondrial intermembrane space (IMS), respectively. The communication with two neighboring modules occurs via stimulation of mitochondrial ATP production by ADP and Pi from the IMS and via time-varying cytosolic ATP hydrolysis during contraction.
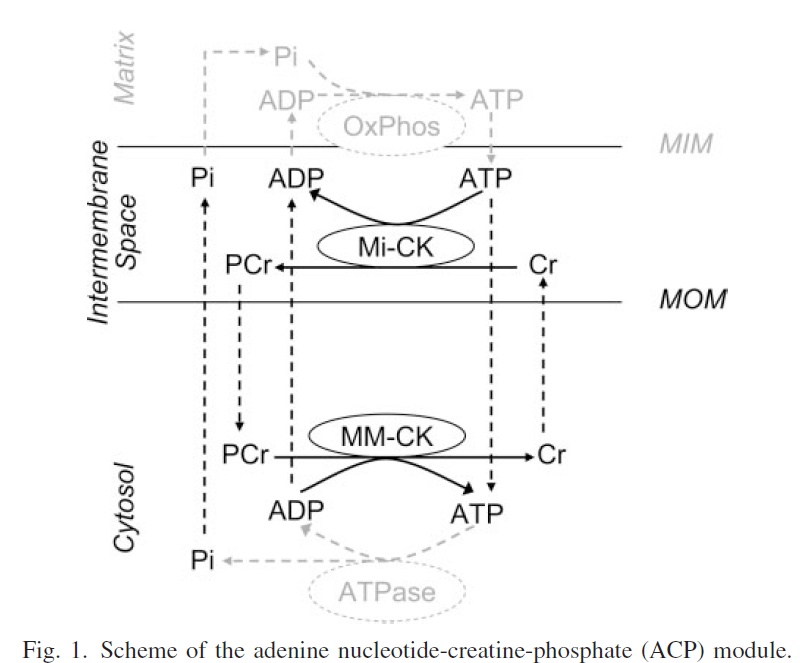
(From vanBeek, 2007)
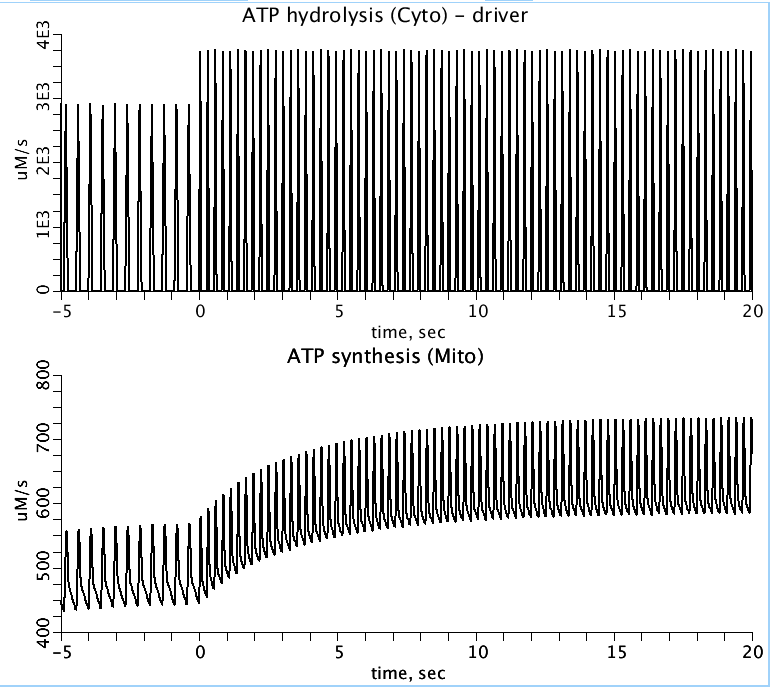
Figure 2: Model reproduction of figure 2 from vanBeek, 2007 paper. Time course of ATP production by the mitochondria in response to a step in heart rate at time t=0. Top, the pulsatile forcing function, which represents ATP hydrolysis during cardiac systole. Bottom, the response of mitochondrial ATP production.
Equations
The equations for this model may be viewed by running the JSim model applet and clicking on the Source tab at the bottom left of JSim's Run Time graphical user interface. The equations are written in JSim's Mathematical Modeling Language (MML). See the Introduction to MML and the MML Reference Manual. Additional documentation for MML can be found by using the search option at the Physiome home page.
- Download JSim model MML code (text):
- Download translated SBML version of model (if available):
- No SBML translation currently available.
- Information on SBML conversion in JSim
We welcome comments and feedback for this model. Please use the button below to send comments:
- (primary) vanBeek JHGM, Adenine nucleotide-creatine-phosphate module in myocardial metabolic system explains fast phase of dynamic regulation of oxidative phosphorylation, Am J Physiol Cell Physiol 293: C815–C829, 2007
- Beard DA. A biophysical model of the mitochondrial respiratory system and oxidative phosphorylation. PLoS Comput Biol 1: e36, 2005.
- Saks VA, Chernousova GB, Gukovsky DE, Smirnov VN, Chazov EI. Studies of energy transport in heart cells. Mitochondrial isoenzyme of creatine phosphokinase: kinetic properties and regulatory action of Mg2+ ions. Eur J Biochem 57: 273–290, 1975
Please cite https://www.imagwiki.nibib.nih.gov/physiome in any publication for which this software is used and send one reprint to the address given below:
The National Simulation Resource, Director J. B. Bassingthwaighte, Department of Bioengineering, University of Washington, Seattle WA 98195-5061.
Model development and archiving support at https://www.imagwiki.nibib.nih.gov/physiome provided by the following grants: NIH U01HL122199 Analyzing the Cardiac Power Grid, 09/15/2015 - 05/31/2020, NIH/NIBIB BE08407 Software Integration, JSim and SBW 6/1/09-5/31/13; NIH/NHLBI T15 HL88516-01 Modeling for Heart, Lung and Blood: From Cell to Organ, 4/1/07-3/31/11; NSF BES-0506477 Adaptive Multi-Scale Model Simulation, 8/15/05-7/31/08; NIH/NHLBI R01 HL073598 Core 3: 3D Imaging and Computer Modeling of the Respiratory Tract, 9/1/04-8/31/09; as well as prior support from NIH/NCRR P41 RR01243 Simulation Resource in Circulatory Mass Transport and Exchange, 12/1/1980-11/30/01 and NIH/NIBIB R01 EB001973 JSim: A Simulation Analysis Platform, 3/1/02-2/28/07.

