Model effect of erythrocyte membrane on exchange of solutes between erythrocytes (red blood cells) and plasma water.
The erythrocyte membrane potentially limits the exchange of solutes between erythrocytes (red blodd cells) and plasma water. The multiple indicator dilution technique can be used to investigate the effect of this barrier on the exchange of tracer substances.
Theory
Single capillary
The concentrations in plasma, Cp and in erythrocytes, CRBC are discribed by the following system of partial differential equations in time t and space x:
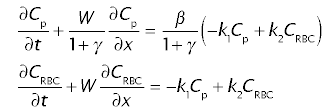
where Cp and CRBC are concentrations in plasma and in erythrocytes, W is the linear velocity of erythrocytes within the sinusoidal lumen, k1 and k2 are transfer coefficients or rate constants, defining the transfer of tracer between plasma and the cytoplasm of perythrocytes, β is the ratio of the red cell water space to the plasma water space in the blood, t0 is the large vessel transit time, and γ is the ratio of the extravascular water space to the sinusoidal plasma water space.
Preequilibrated injection
Tracer to be injected are added to a blood sample or the same hematocrit as in systemic blood some time before injection. In this case, the tracer will equilibrate between plasma and erythrocytes, and the boundary conditions are
- at t = 0: Cp = 0 and CRBC = 0;
- at x = 0: Cp = (1 + β)/(1 + k1/k2 β) Cin and CRBC = k1/k2Cp
where Cin is the tracer concentration at the inflow of the organ. In order to make calculations easier, concentrations are normalized by dividing through the injected amount, and distances are normalized by dividung through the linear velocity W, thus assuiming W = 1.
Whole organ
The concentration in the blood leaving the organ through the portal vein is calculated as the average weighted according to the flow in all sinusoidal paths (analogous to the case of "barrier-limited transport") and the relative volume of plasma and erythrocytes:
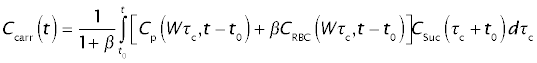
Experimental design
The following tracers were injected into the portal vein of an anesthetized dog:
- Erythrocytes (Ery) labeled with 51Cr
- Labeled thiourea (NH2-14CS-NH2), a substance that penetrates easily into hepatocytes, and less easily into erythrocytes
- Tritiated water(3HOH)
Samples from a hepatic vein were analyzed for beta radioactivity, and the various isotopes where distinguished by scintillation spectrometry. In the following applet o click on "Run" to calculate the curves o click on plotpage_1 to see the data and the calculated curves
Calculations
Parameter sets
There are two parameter sets in this JSim model project file:
- HiHct: High hematocrit (0.42)
- LowHct: The hematocrit was lowered to 0.25 by bleeding and plasma substitution
To change the parameter set:
- Select Load project parameter set from the ParSet pulldown menu
- Choose the desired parameter set. This will automatically change the paremters as well as the data for the reference curve.
- Click on "Run" to use the new parameter set.
- On the plotpage, select number 1 from the "Curve" pulldown menu
- Select data set from the "Data" pulldown menu
- Repeat the same procedure for plot number 2.
Optimization
If you want to optimize the parameters of the newly selected experiments
- Click on the Optimizer tab (at the bottom of the left pane)
- Change all the DataSet entries under the "Data to Match" heading.
- Click on the Dataset entries to get a selection of data sets to choose from.
- Click on the Curve entries to get a selection of data columns to choose from.
- Hit the "Run" button.
We welcome comments and feedback for this model. Please use the button below to send comments:
CA Goresky, GG Bach and BE Nadeau. Red cell carriage of label: its limiting effect on the exchange of materials in the liver. Circulation Research:36, 328-351, 1975
Author: Andreas J. Schwab (andreas.schwab@mcgill.ca)
Please cite https://www.imagwiki.nibib.nih.gov/physiome in any publication for which this software is used and send one reprint to the address given below:
The National Simulation Resource, Director J. B. Bassingthwaighte, Department of Bioengineering, University of Washington, Seattle WA 98195-5061.
Model development and archiving support at https://www.imagwiki.nibib.nih.gov/physiome provided by the following grants: NIH U01HL122199 Analyzing the Cardiac Power Grid, 09/15/2015 - 05/31/2020, NIH/NIBIB BE08407 Software Integration, JSim and SBW 6/1/09-5/31/13; NIH/NHLBI T15 HL88516-01 Modeling for Heart, Lung and Blood: From Cell to Organ, 4/1/07-3/31/11; NSF BES-0506477 Adaptive Multi-Scale Model Simulation, 8/15/05-7/31/08; NIH/NHLBI R01 HL073598 Core 3: 3D Imaging and Computer Modeling of the Respiratory Tract, 9/1/04-8/31/09; as well as prior support from NIH/NCRR P41 RR01243 Simulation Resource in Circulatory Mass Transport and Exchange, 12/1/1980-11/30/01 and NIH/NIBIB R01 EB001973 JSim: A Simulation Analysis Platform, 3/1/02-2/28/07.

