A model for triple-labeled indicator dilution experiments in a four region tissue exchange model.
Further reading: Distributed Blood Tissue Exchange Models Explained
Description
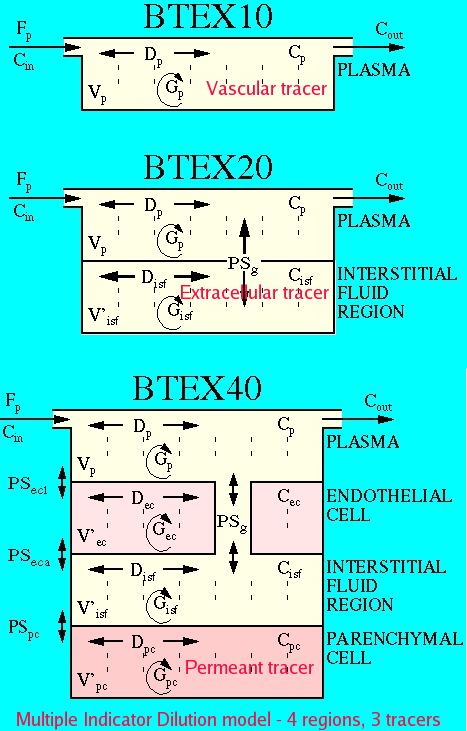
This is a model for triple-labeled indicator dilution experiments in a four region tissue exchange model. A BTEX10 model is used for the vascular tracer (plasma), a BTEX20 model is used for the extracellular tracer (plasma and interstitial fluid region), and a BTEX40 model is used for the permeant tracer (plasma ,interstitial fluid region, and endothelial and parenchymal cell regions. Extraction is calculated and used to calculate estimated permeability-surface area factors between various regions. See MID4ode for a version using compartment models
Equations
Vascular Equations
Differential Equation
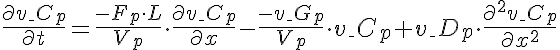
Left Boundary Condition
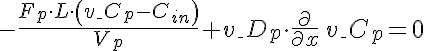
Right Boundary Conditions
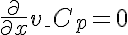 ,
, 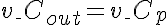
Initial Conditions
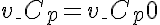 or
or 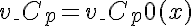
Extracellular Equations
Differential Equations
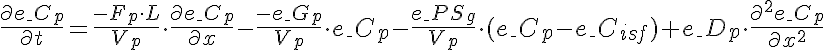
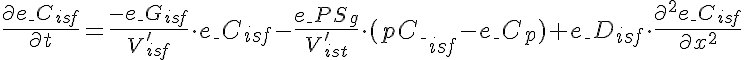
Left Boundary Conditions
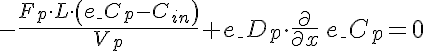 ,
, 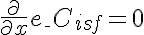
Right Boundary Conditions
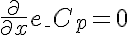 ,
, 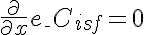 ,
, 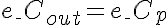
Initial Conditions
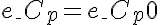 ,
, 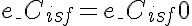 or
or 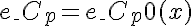
Permeant Equations
Differential Equations
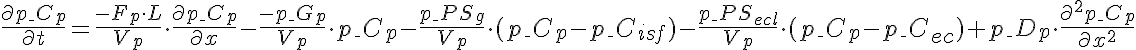
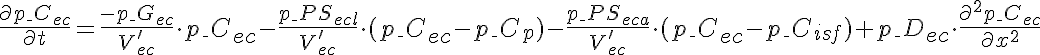
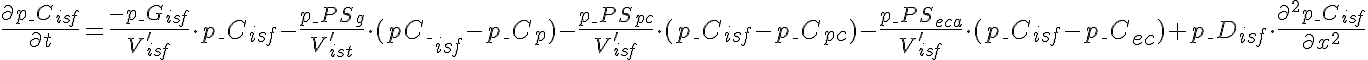
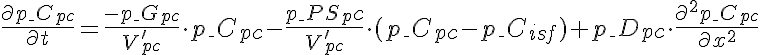
Left Boundary Conditions
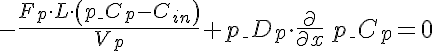 ,
, 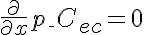 ,
, 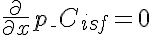 ,
, 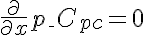 .
.
Right Boundary Conditions
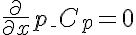 ,
, 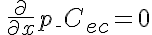 ,
, 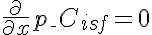 ,
, 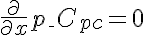 ,
, 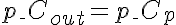 .
.
Initial Conditions
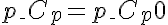 ,
, 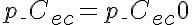 ,
, 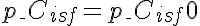 ,
, 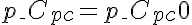 or
or 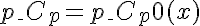 , , , .
, , , .
The equations for this model may be viewed by running the JSim model applet and clicking on the Source tab at the bottom left of JSim's Run Time graphical user interface. The equations are written in JSim's Mathematical Modeling Language (MML). See the Introduction to MML and the MML Reference Manual. Additional documentation for MML can be found by using the search option at the Physiome home page.
- Download JSim model MML code (text):
- Download translated SBML version of model (if available):
- No SBML translation currently available.
- Information on SBML conversion in JSim
We welcome comments and feedback for this model. Please use the button below to send comments:
W.C. Sangren and C.W. Sheppard. A mathematical derivation of the exchange of a labelled substance between a liquid flowing in a vessel and an external compartment. Bull Math BioPhys, 15, 387-394, 1953. C.A. Goresky, W.H. Ziegler, and G.G. Bach. Capillary exchange modeling: Barrier-limited and flow-limited distribution. Circ Res 27: 739-764, 1970. J.B. Bassingthwaighte. A concurrent flow model for extraction during transcapillary passage. Circ Res 35:483-503, 1974. B. Guller, T. Yipintsoi, A.L. Orvis, and J.B. Bassingthwaighte. Myocardial sodium extraction at varied coronary flows in the dog: Estimation of capillary permeability by residue and outflow detection. Circ Res 37: 359-378, 1975. C.P. Rose, C.A. Goresky, and G.G. Bach. The capillary and sarcolemmal barriers in the heart--an exploration of labelled water permeability. Circ Res 41: 515, 1977. J.B. Bassingthwaighte, C.Y. Wang, and I.S. Chan. Blood-tissue exchange via transport and transformation by endothelial cells. Circ. Res. 65:997-1020, 1989. Poulain CA, Finlayson BA, Bassingthwaighte JB.,Efficient numerical methods for nonlinear-facilitated transport and exchange in a blood-tissue exchange unit, Ann Biomed Eng. 1997 May-Jun;25(3):547-64.
Please cite https://www.imagwiki.nibib.nih.gov/physiome in any publication for which this software is used and send one reprint to the address given below:
The National Simulation Resource, Director J. B. Bassingthwaighte, Department of Bioengineering, University of Washington, Seattle WA 98195-5061.
Model development and archiving support at https://www.imagwiki.nibib.nih.gov/physiome provided by the following grants: NIH U01HL122199 Analyzing the Cardiac Power Grid, 09/15/2015 - 05/31/2020, NIH/NIBIB BE08407 Software Integration, JSim and SBW 6/1/09-5/31/13; NIH/NHLBI T15 HL88516-01 Modeling for Heart, Lung and Blood: From Cell to Organ, 4/1/07-3/31/11; NSF BES-0506477 Adaptive Multi-Scale Model Simulation, 8/15/05-7/31/08; NIH/NHLBI R01 HL073598 Core 3: 3D Imaging and Computer Modeling of the Respiratory Tract, 9/1/04-8/31/09; as well as prior support from NIH/NCRR P41 RR01243 Simulation Resource in Circulatory Mass Transport and Exchange, 12/1/1980-11/30/01 and NIH/NIBIB R01 EB001973 JSim: A Simulation Analysis Platform, 3/1/02-2/28/07.

