Contents:
- Optimization Overview
- JSim Optimization Example
- Parameter Indentifiability
- Parameter Selection
- Data Selection
- Algorithms
- Stopping Criteria
- Monte Carlo Analysis
Optimization Overview
JSim optimization is the process of improving model fits to experimental data through the adjustment of parameters. Parameter adjustment may be performed manually or automatically. In the manual case, the modeler uses mathematical or physiological insight to adjust parameter values by informed trial-and-error. Evaluation of the optimization is usually done by examining the visual fit of model ouputs to experimental data, although it may also be informed by statistical measures such as root-mean-square(RMS) error.
Also, see Introduction to the JSim Optimizers for an initial exploration of optimizing model parameters.
In the automated case, the modeler sets up an optimization run and lets JSim's internal optimization algorithms do the desired parameter adjustment. Optimization run specifications can get fairly complex but, at minimum, the modeler must specify the parameter(s) to vary, the experimental reference data and the model outputs that will be compared to it. With this information, the optimizer will run the model multiple times. After each run, the RMS error between the experimental data and model outputs is calculated, and used as a basis for subsequent adjustment to the specified parameters. The goal of this parameter adjustment is to reduce RMS error ideally to zero (indicating a "perfect" model fit), but more practically to some acceptably small positive value (indicating an "acceptable" model fit).
RMS error may be thought of as a scalar function of (perhaps multi-dimensional) parameter space, and optimization may be thought of as finding the point in parameter space corresponding to minimum RMS error. In this view, the RMS error function is a topographical map, with the desired optimization minimum corresponding to the deepest valley in the terrain. (We will limit our discussion here to deterministic models, where a given parameter set always produces the same outputs and same RMS error. Optimization of stochastic models, in which model outputs may vary from one run to another, even with the same set of parameters, is, obviously, a more complicated matter.) Several possible problems are worth noting. First, there may be no unique minimum value for RMS error, which may be unbounded (e.g. RMS-error = exp(-P)), or have multiple identical minima (e.g. RMS-error = 1 + sin(P)). Second, our knowledge of the RMS error function is usually limited to a finite set of points sampled via model runs. This limited sampling means it is quite possible that major topographical features of the RMS error function will be missed - that is, we may find a "local minimum" but not a "global minimum".
FIGURE: GRAPHS OF MINIMIZATION ISSUES
ALGORITHMS: JSim provides 8 differenct algorithms for optimization: Simplex, GGopt, Neldermead, NL2SOL, Sensop, Simanneal, Genetic and GridSearch. A detailed description of these algorithms will be described in a later section of this tutorial. The first five of these algorithms are considered "steepest descent" algorithms, in that they attempt to trace a path of maximum gradient down the RMS error function map. Steepest descent algorithms work very efficiently, but since the descent is based on the initial parameter estimate, they can easily get stuck in local minima if the initial parameter estimate is not close enough to the global minimal value. In contrast, the last three algorithms (Simanneal, Genetic and GridSearch) use various strategies to sample a wider set of values in parameter space. This has the advantage of being less likely to get stuck in local minima, but with the disadvantage of requiring many more model runs, and thus more compute time.
PARAMETER BOUNDS: One or more parameters may be specified for optimization during a single optimization run. When the algorithm used is Simplex, Sensop, SimAnneal, Genetic or GridSearch, the parameters must be bounded, meaning that a minimum and maximum value must be specified for each parameter. During the optimization run, no value outside the specified range will be used for a model run. In remaining algorithms (GGopt, NelderMead and NL2SOL), the parameters are unbounded. No minimum or maximum values need be specified and parameter values could conceivable assume any real value. Unbounded parameters can be problematic because model equations may make sense only for certain real values. For example, a chemical concentration can never go negative. Using unbounded parameters when optimizing a concentration parameter could cause the algorithm to test a negative concentration value, causing the model run, and thus the optimization run, to fail. For this reason, you should be careful to understand the stability characteristics of a parameter over the entire range of real values if you select an unbounded optimizer algorithm.
EXAMPLE NEEDED HERE OF MODEL NEEDING BOUNDED ALGORITHM
JSim Optimizer Example
Suppose you wish to match output from a given model to data taken from a scientific experiment. The general process is as follows:
1) Create a data file with the experimental results and import it into the project that contains your model (see Data Files and Project Data Sets ).
2) Decide which model outputs should best match which data curves. Typically this involves a bit of exploration of model parameter space using plot pages.
3) Configure the model's "Optimizer" sub-tab with the parameters you wish to vary and the model outputs and data curves which should ideally match.
4) Push the optimizer's "Run" button and watch as the optimizer works.
5) Examine optimizer results and model outputs at new optimized parameter values.
Starting the tutorial
on your computer:
- If you have not already done so, download and install JSim on your computer.
- , the model file for this tutorial.
- , the data file for this tutorial.
- Start JSim on your computer
- Select "Import model file..." from the "Project" tab "Add" menu and load the opt1.mod model file.
- Select "Import data file..." from the "Project" tab "Add" menu and load the optdata1.tac data file.
in the JSim applet:
Understanding the model
Examine the model MML to see how it works. Compile and run the model and plot the output variable named "u", which represents exponential decay. u's values are controlled by the parameters "amp" and "decay", both of which have 1 as a default value. Run the model with various values of amp and decay to get a feeling for how they affect the affect u. When you are done, restore amp and decay to their default values.
Now superimpose the curve R1s1 from the data set optdata to the plot containing u. Alter plot colors or other characteristics so that you can clearly distinguish between model output and fixed data. Now run the model varying amp and decay until you get a reasonable match between u and R1s1. This is the process the optimizer will automate. Note that both amp and decay must be altered from their defaults to get a reasonable fit. If you are confused about any of the above operations, you should probably review the documents mentioned in the introduction.
A Sample Optimization
We will now setup the optimizer to automatically perform the optimization you did "by hand" in the previous section. Restore amp and decay to their default values, and then select expon's "Optimizer" sub-tab which configures an optimization run.
The configuration is divided vertically into three sections. We'll ignore the top section for now. Look at the "Parameters to Vary" section. Here is where we will specify that amp and decay are the parameters we wish to vary. Enter "amp" (no quotes) in the Parameter column or select amp from a parameter list by clicking the down arrow button to the right of the box. The line will blacken showing the current value of amp (which should be 1) in the Start column. Fill in 0.5 and 5 in the Min and Max columns to indicate the minimum and maximum values of amp you wish to optimizer to consider. The Step column prescribes in the initial absolute (not relative) change in parameter value. For this tutorial, the default value is adequate. Once all fields in the line have contents, a check box will appear in the OK column. Optimization of this parameter may be temporarily disabled by unchecking the box. A question mark will appear in the OK column, if there is some problem with the line. Clicking on the question mark will display a message telling what the problem is.
A second (blank) line will have opened when you entered amp in the first line. Specify the parameter decay in the second line as you did amp in the first. Use the same min and max values. The "Parameters to Vary" section should now show two blackened lines, and a third blank one.
Now look at the "Data to Match" section. Here we will tell the optimizer to get the best match it can between data curve R1s1 and model output u. The first column is the dataset from which to draw the data curve. Since your project contains only a single data set (optdata), it will be automatically filled in. If you use the optimizer in projects with more than one data set, you will need to select which data set to use. In the "Curve" column enter "R1s1" no quotes or select it from the popdown list. The box should blacken. In the "Par/Expr" column, enter "u" (no quotes) or select it from the pop-down list. The first line under "Data to Match" should now be completely blackened and a check box should appear in the OK column. This checkbox (or the associated question mark) function as their counterparts in the "Parameters to Vary" section. Ignore the "Pwgt" and "Cwgt" columns for now.
The optimizer is now ready to be run. Press the "Run" button in the optimizer configuration menubar. The optimizer works by running the model for various parameter values, guessing after each run other values to try. In our case the model runs very quickly, so entire optimization run should only take a few seconds. Your plot page should now show good agreement between model output and reference data. The optimized values for amp and decay are now shown in the "Start" column of the "Parameters to Vary" section. Note that the values are good, but not perfect. Perfect values would be amp=2 and decay=3.
Graphs and Reports
The "Optimizer" sub-tab, has three sub-sub-tabs along the top margin. The "Config" (sub-sub-)tab was used to configure the optimization run. Useful results from the most recent optimization run can be viewed in the "Graph" and "Report" tabs. The "Graph" tab has seven different "View" options which should be fairly self-explanatory after you see them once. The "Report" tab should also be easy to understand. Take a few minutes to examine the graphs and report. Note that since we did no relative data weighting, the weighted and unweighted residual graphs are identical and the "Point weigths" graph contains nothing useful.
All three Optimizer sub-sub-tabs possess and "Run" button in the menubar. The results of the optimizer will be same in each case. What will vary will be the reporting the user receives as the optimizer works. Currently JSim does not support the user switching between different types of feedback while the optimizer is running.
Complications
Real world models and data will not fit so easily as in this idealized example. This introduction serves only to show how to operate the JSim optimizer, but does attempt to address the large scientific problem of how best to optimize parameters to reference data. Future tutorials will address these issues. Some complicating factors are:
- Errors in mathematical formulation of models which no mere parameter adjustments can hope to compensate for.
- Noisy data may good fits difficult. Models may be expected to fit multiple data curve. Relative weighting of RMS error between data points or data curves may be highly subjective.
- Optimizer algorithms generally perform more and more poorly the larger the number of varying parameters.
Parameter Indentifiability
A model parameter is identifiable if it's value may be discerned from enough observations of model output. Consider the model:
See ident.proj
math main {
realDomain t;
t.min=0; t.max=1; t.delta=.1;
real a = 1;
real b = 1;
real c(t) = a*exp(-t);
real d(t) = a/b*exp(-t);
}
Output c(t) has no dependence on parameter b, so an optimization run varying only parameter b, and matching output c(t) against some reference curve R, could not possibly return a reasonable value of b. Since c(t) does depend on a, a reasonable result.
EXAMPLE HERE: OPTIM b, c(t)
Also note, that output d(t) is dependent on the ratio of a and b, not their absolute values. As a result, an optimization run varying parameters a and b, and matching output d(t) against some reference curve R, may return for example, doubling both a and b would result in that same values for c(t). This makes it impossible to identify both a and b using information from c(t). If we configure a JSim optimization run to vary parameters a and b, matching output d(t) against a data curve R = 2*exp(-t), a "perfect" fit could be obtained, but the optimized values of (a,b) might be any number of different values e.g. (2,1), (4,2), (.5, .25). JSim's return a particular (a,b) pair from the optimization indicates only Incautious interpretation of the optimization results might but only a single value is returned.
Using the covariance matrix
TERMINATION: JSim optimization runs may be terminated in several ways. The control parameter
EVALUATION: Supposing Covariance matrix;
examine model misfit of data in plot
choose model parameters to optimize
choose data to optimize on
run optimization
examine improved model fit of data in plot
examine results graphs (esp. residuals)
examine optimization report
note covariance matrix which will be examined in detail later in tutorial
Parameter Selection
Independence
Covariance Matrix
meaning of use to eliminate some parameters from optimization possible rewriting of model to remove covariant parameters
Sensitivity Analysis
Data selection
See Importing data sets into JSim for information about formatting of data for use within JSim.
Project data sets
Point weighting
Curve weighting
Multiple curves to match
Algorithms (strengths/weaknesses of each algorithm + tuning parameters)
Some terminology is useful when discussing the merits of optimization algorithms:
- Bounded algorithms are those that require upper and lower bounds for each parameter varied. Unbounded algorithms require no such bounds.
- Parallel algorithms are those that can take advantage of multiple system processors for faster processing. See Using JSim Multiprocessing for further information on JSim multiprocessing.
All of JSim's currently available optimization algorithms share the following control parameters:
- max # runs: The optimizer will stop if it has run the model this many times.
- min RMS error: The optimizer will stop if the mean RMS error between reference data and model output is less than this value.
Algorithm-specific control parameters are listed with each algorithm description.
Simplex
JSim's simplex is a bounded, non-linear steepest-descent algorithm. This algorithm does not currently support parallel processing. (description needs work)
Algorithm-specific control parameters:
- parameter initial step: default=0.01
- minimum par step: The optimizer will stop if it is considering a parameter step value that vary less than this value.
GGopt
GGopt is an unbounded non-linear algorithm originally written by Glad and Goldstein. This algorithm does not currently support parallel processing. (description needs work)
Algorithm-specific control parameters:
- minimum par step: The optimizer will stop if it is considering a parameter step value that vary less than this value.
- maximum # iterations: default=10
- minimum gradient: default=1e-6
- relative error: default=1e-6
Nelder-Mead
Nelder-Mead is an unbounded, steepest descent algorithm by Nelder and Mead. It is also called non-linear Simplex. This algorithm supports multiprocessing (MP).
During each iteration in a P-parameter optimization, this algorithm performs a P or P+1 parmeter queries (model runs). Several additional single queries are also performed. Ideal MP speedup on an N-processor system on be roughly of order P (if P is a factor of N), or order N (if N is a factor of P).
This algorithm differs from the previously available JSim "Simplex" (above) in that:
- it is unbounded, while "Simplex" is bounded;
- it supports MP, while "Simplex" does not;
- it is a newer implementation of the algorithm (Java vs. Fortran).
Algorithm-specific control parameters:
- parameter initial step: default=0.01
- minimum par step: The optimizer will stop if it is considering a parameter step value that vary less than this value.
GridSearch
GridSearch is a bounded, parallel algorithm. The algorithm operates via progressively restricted search of parameter space on a regularly spaced grid of npoints per dimension. Each iteration, npoints^nparm points are searched for the minimum residual. Each parameter dimension is then restricted to one grid delta around that minimum and the search repeats until stopping criteria are met.
Search bounds in each dimension narrow by a factor of at least 2/(npoints-1) each iteration. Thus, npoints must be at least 4. Each iteration requires up to npoints^nparm residual calculations. Residual calculations are reused when possible, and this reuse is most efficient when npoints is 1 + 2^N for some N. Therefore, npoints defaults to 5, which is the smallest "efficient" value.
This algorithm is very not efficient for very smooth residual functions in high-dimensional space. It works well on noisy functions when low accuraccy situations (e.g. 3 significant digits required). With npoints large, it copes well with multiple local minima. An effective strategy may be to use several interations of GridSearch to estimate a global minimum, and then use a steepest-descent algorithm to fine tune the answer.
The number of points searched during each iteration is typically large compared to the number of available processors. Typical MP speedup on an N-processor system is therefore on the order of N.
Algorithm-specific control parameters:
- minimum par step: The optimizer will stop if it is considering a parameter step value that vary less than this value.
- max # iterations: stop after this many iterations, default=10.
- # grid points: npoints above, default=5.
Nl2sol
This version of Nl2sol is derived from NL2SOL, a library of FORTRAN routines which implement an adaptive nonlinear least-squares algorithm. It has been modified to perform all calculations in double precision. It is an unbounded optimizer. This optimizer does not support multi-processing.
Algorithm-specific control parameters:
- maximum # runs: default=50
- relative error: default=1e-6
Sensop
Sensop is a variant of Levenberg-Marquardt algorithm that utilized the maximum parameter sensitivity to determine step size. It is a bounded optimizer, supporting multiprocessing.
Algorithm-specific control parameters:
- minimum gradient: default=50
- minimum par step: The optimizer will stop if it is considering a parameter step value that vary less than this value.
Simulated Annealing
Simulated annealing is an algorithm inspired by the annealing process in metullurgy. As the problem "cools" from its initial "temperature", random fluctuations of model parameters are reduced. JSim implements a bounded version of this algorithm that supports multiprocessing.
Algorithm-specific control parameters:
- initial temperature:default=100. This parameter has no physical meaning (e.g. Kelvin), but must be positive.
Genetic Algorithms
This algorithm is available only in JSim version 2.01 and above.
Genetic algorithms are a family of algorithms that generate a population of candidate solutions and then select the best solutions in each iteration. A new population of solutions is created during each iteration. There are different ways of specifying how a new population is generated from the existing population. The error calculation is used to score and rank the candidate solution. The "fit" individuals in the existing population are selected using one of three methods: (1) roulette-wheel, (2) tournament selection, or (3) elitism. In the roulette-wheel method, the probability of a solution being selected is inversely proportional to the error. In the tournament selection, two random solutions are selected and the one with the lower error is placed in the new population. In elitism, all the solutions with the lowest errors (with a cutoff fraction) are selected. New solutions are selected by "mutating" and "crossing over" existing solutions.
Algorithm-specific control parameters:
- Population:default= 25. Number of trials in each generation. If max # runs < Population then Population is defaulted to the value of max # runs and only the parent generation is calculated. Suggested values are max # runs = 400, Population=100 for four generations.
- Mutation rate:default = 0.1. The probability of any single parameter getting changed.
- Crossover rate:default = 0.5. The probability of creating a new solution by combining existing solutions.
- Select Method: default= 1. Acceptable values are (1) roulette-wheel, (2) tournament selection, and (3) elitism. See above description identifying these terms.
- Mutation step:default=0.05. The amount by which a mutation changes a parameter, specified as a fraction of the range.
- Elite cutoff:default=0.5. Fraction of the population considered fit for the elitism selection method (Select Method=3.)
Stopping Criteria
manual termination when "good enough"
Some stopping criteria are common to all algorithms, while some are algorithm-specific. Common stopping criteria are described below. See JSim Optimization Algorithms for algorithm-specific details.
The common stopping criteria are:
- Max # runs: The optimizer will stop if it has run the model this many times.
- Min RMS error: The optimizer will stop if the mean RMS error between reference data and model output is less than this value. The RMS error is calculated as
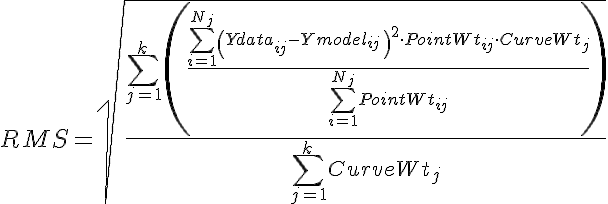
where there are j=1 to k curves, each curve having a curve weight, CurveWeightj, and having i=1 to Nj data points, Ymodelij, and corresponding point weights, PointWtij.
- Min par step: The optimizer will stop if it is considering parameter values that vary less than this value.
If unrealistic values for the stopping criteria are specified, the model optimizer may either run forever, or stop before any useful work is done. You always have the option of cancelling a long optimization run, and the best values so far calculated will be retained.
Monte Carlo Analysis
Use the following links to learn how to use JSim's Monte Carlo analysis tool:
- Introduction to JSim Monte Carlo analysis.
- Simple Monte Carlo analysis
- Correlation of parameters: sensitivity of a given set of parameters versus multiple sets of parameter estimates (Monte Carlo approach).
- Parameter optimization of a three exponential curve, compare and contrast.
- Aspirin clearance: Monte Carlo analysis used in a model analysis.
Comments or Questions?
Model development and archiving support at https://www.imagwiki.nibib.nih.gov/physiome provided by the following grants: NIH U01HL122199 Analyzing the Cardiac Power Grid, 09/15/2015 - 05/31/2020, NIH/NIBIB BE08407 Software Integration, JSim and SBW 6/1/09-5/31/13; NIH/NHLBI T15 HL88516-01 Modeling for Heart, Lung and Blood: From Cell to Organ, 4/1/07-3/31/11; NSF BES-0506477 Adaptive Multi-Scale Model Simulation, 8/15/05-7/31/08; NIH/NHLBI R01 HL073598 Core 3: 3D Imaging and Computer Modeling of the Respiratory Tract, 9/1/04-8/31/09; as well as prior support from NIH/NCRR P41 RR01243 Simulation Resource in Circulatory Mass Transport and Exchange, 12/1/1980-11/30/01 and NIH/NIBIB R01 EB001973 JSim: A Simulation Analysis Platform, 3/1/02-2/28/07.

