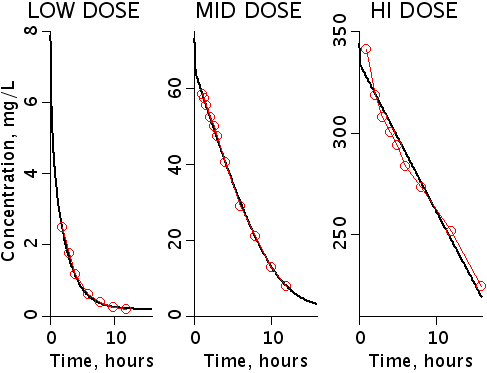
Salicylic acid (SA) clearance for three different dose ranges is modeled as an enzyme reaction. Model parameters are optimized and Monte-Carlo analysis is performed to robustly quantify parameter estimation, variance and covariance with other parameters in the presence of noisy data..
Description
This is an enzyme conversion model for salicylic acid clearance
simultaneously fitting three independent data sets using shared parameters.
The initial concentrations of salicylic acid range over almost two orders of
magnitude.
kon1-> koff2-> Gp->
LSA+LE<-----> LSAE <-----> LE+LP-----> (Low Dose)
<-koff1 <-kon2
kon1-> koff2-> Gp->
MSA+ME<-----> MSAE <-----> ME+MP-----> (Mid Dose)
<-koff1 <-kon2
kon1-> koff2-> Gp->
HSA+HE<-----> HSAE <-----> HE+HP-----> (Hi Dose)
<-koff1 <-kon2
koff1=KD1*kon1 koff2=KD2*kon1
where LSA, MSA, and HSA are Salicylic Acid concentrations,
HE, ME, and HE are the free enzyme concentrations,
LSAE, MSAE, and HSAE are salicylic acid-enzyme complex concentrations, and
LP, MP, and HP are the product concentrations.
The parameters kon1, koff2, koff1, kon2, Gp, and the total amount of enzyme
are the same for all three models.
Equations
Algebraic Equations
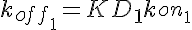
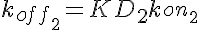
Ordinary Differential and Mass Balance Equations
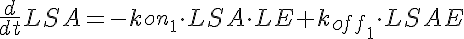
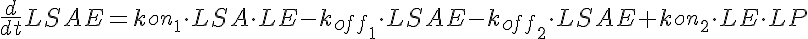
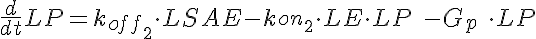
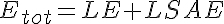
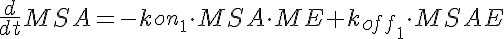
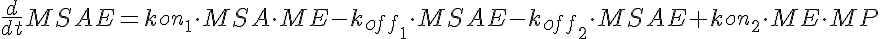
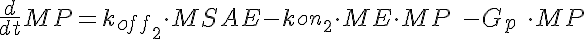
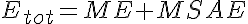
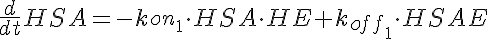
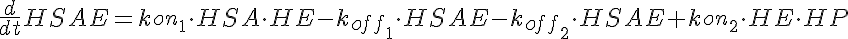
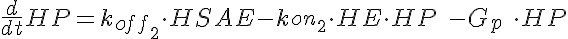
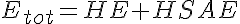
Initial Conditions
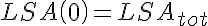 ,
, 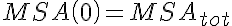 ,
, 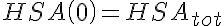 .
.
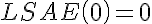 ,
, 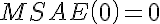 ,
, 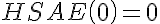 .
.
The equations for this model may be viewed by running the JSim model applet and clicking on the Source tab at the bottom left of JSim's Run Time graphical user interface. The equations are written in JSim's Mathematical Modeling Language (MML). See the Introduction to MML and the MML Reference Manual. Additional documentation for MML can be found by using the search option at the Physiome home page.
- Download JSim model MML code (text):
- Download translated SBML version of model (if available):
We welcome comments and feedback for this model. Please use the button below to send comments:
IH Benedek, AS Joshi, HJ Pieniaszek, SY King and DM Kornhauser; Variability in the pharmacokinetics and pharmacodynamics of low dose aspirin in healthy male volunteers. J. Clin. Pharmacol 35: 1181-1186, 1995. L Aarons, K Hopkins, M Rowland, S Brossel, and JF Thiercelin: Route of administration and sex differences in the pharmacokinetics of aspirin, administered as its lysine salt. Pharmaceutical Res 6: 660-666, 1989. LF Prescott, M Balali-Mood, JA Critchley, AF Johnstone, AT Proudfoot; Diuresis or urinary alkalinisation for salicylate poisoning?. Br Med J, 285 1383-1384, 1982.
Please cite https://www.imagwiki.nibib.nih.gov/physiome in any publication for which this software is used and send one reprint to the address given below:
The National Simulation Resource, Director J. B. Bassingthwaighte, Department of Bioengineering, University of Washington, Seattle WA 98195-5061.
Model development and archiving support at https://www.imagwiki.nibib.nih.gov/physiome provided by the following grants: NIH U01HL122199 Analyzing the Cardiac Power Grid, 09/15/2015 - 05/31/2020, NIH/NIBIB BE08407 Software Integration, JSim and SBW 6/1/09-5/31/13; NIH/NHLBI T15 HL88516-01 Modeling for Heart, Lung and Blood: From Cell to Organ, 4/1/07-3/31/11; NSF BES-0506477 Adaptive Multi-Scale Model Simulation, 8/15/05-7/31/08; NIH/NHLBI R01 HL073598 Core 3: 3D Imaging and Computer Modeling of the Respiratory Tract, 9/1/04-8/31/09; as well as prior support from NIH/NCRR P41 RR01243 Simulation Resource in Circulatory Mass Transport and Exchange, 12/1/1980-11/30/01 and NIH/NIBIB R01 EB001973 JSim: A Simulation Analysis Platform, 3/1/02-2/28/07.

