(Matlab) Metabolic Dynamics in Skeletal Muscle during Acute Reduction in Blood Flow and Oxygen Supply to Mitochondria: In-Silico Studies Using a Multi-Scale, Top-Down Integrated Model.
Description
(Abstract) Control mechanisms of cellular metabolism and energetics in skeletal muscle that may become evident in response to physiological stresses such as reduction in blood flow and oxygen supply to mitochondria can be quantitatively understood using a multi-scale computational model. The analysis of dynamic responses from such a model can provide insights into mechanisms of metabolic regulation that may not be evident from experimental studies. For the purpose, a physiologically based, multi-scale computational model of skeletal muscle cellular metabolism and energetics was developed to describe dynamic responses of key chemical species and reaction fluxes to muscle ischemia. The model, which incorporates key transport and metabolic processes and subcellular compartmentalization, is based on dynamic mass balances of 30 chemical species in both capillary blood and tissue cells (cytosol and mitochondria) domains. The reaction fluxes in cytosol and mitochondria are expressed in terms of a general phenomenological Michaelis-Menten equation involving the compartmentalized energy controller ratios ATP/ADP and NADH/NAD+. The unknown transport and reaction parameters in the model are estimated simultaneously by minimizing the differences between available in vivo experimental data on muscle ischemia and corresponding model outputs in coupled with the resting linear flux balance constraints using a robust, nonlinear, constrained-based, reduced gradient optimization algorithm. With the optimal parameter values, the model is able to simulate dynamic responses to reduced blood flow and oxygen supply to mitochondria associated with muscle ischemia of several key metabolite concentrations and metabolic fluxes in the subcellular cytosolic and mitochondrial compartments, some that can be measured and others that can not be measured with the current experimental techniques. The model can be applied to test complex hypotheses involving dynamic regulation of cellular metabolism and energetics in skeletal muscle during physiological stresses such as ischemia, hypoxia, and exercise.
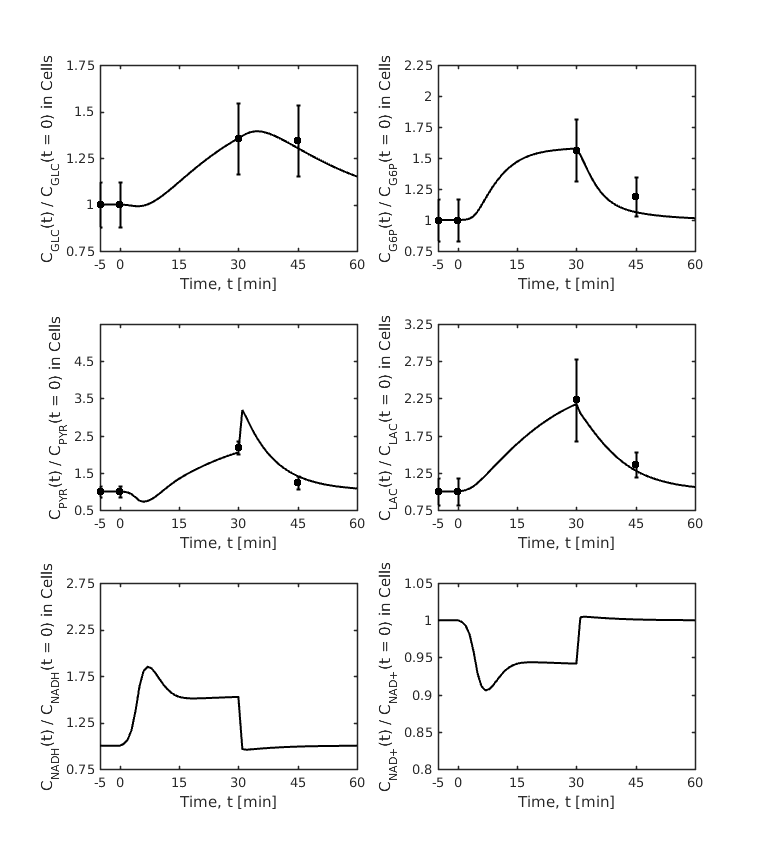
Figure: (from fig 3, Dash et al. 2008) Model-predicted dynamic responses of glycolytic metabolite concentrations and redox states in the muscle tissue cells during the resting, ischemia and recovery periods with varying levels of blood flow reduction and their comparison to the experimental data of Katz 1988. The responses were computed using the estimated optimal parameter values with the ischemia protocol of 25 to 0 min of resting, 0 to 30 min of ischemia, and 30 to 60 min of recovery. The muscle blood flow Q is reduced as a step from 0.9 L/min at rest to Qisch = 0.216 L/min at the onset of ischemia and returned to 0.9 L/min at the onset of recovery. The lines represent the model simulation results with the symbols representing the experimental data points (mean +-SD) corresponding to Qisch = 0.216 L/min (~76% blood flow reduction). The metabolites concentrations are shown in normalized form, normalized with respect to the resting metabolites concentrations. The concentrations in the tissue cells are calculated based on the formula: Ccl = (VcytCcyt+VmitCmit)/Vcl
Equations
The equations for this model may be viewed by opening up the Matlab model files downloaded from below.
Download Matlab model and associated files.
- Run Matlab simulation through 'SkelMusMetab_Driver.m' file after extracting zip file.
We welcome comments and feedback for this model. Please use the button below to send comments:
- (Primary) Dash RK, Li Y, Kim J, Beard DA, Saidel GM, et al. (2008) Metabolic Dynamics in Skeletal Muscle during Acute Reduction in Blood Flow and Oxygen Supply to Mitochondria: In-Silico Studies Using a Multi-Scale, Top-Down Integrated Model. PLoS ONE 3(9): e3168. doi:10.1371/journal.pone.0003168
- Dash RK, Li Y, Kim J, Saidel GM, Cabrera ME (2008) Modeling cellular metabolism and energetics in skeletal muscle: large-scale parameter estimation and sensitivity analysis. IEEE Trans Biomed Eng 55: 1298–1318.
- Cabrera M, Saidel G, Kalhan S (1998) Modeling metabolic dynamics from cellular processes to organ and whole body responses. Prog Biophys Molec Biol 69: 539–557.
- Katz A (1988) G-1,6-P2, glycolysis, and energy metabolism during circulatory occlusion in human skeletal muscle. Am J Physiol Cell Physiol 255: C140–C144.
Please cite https://www.imagwiki.nibib.nih.gov/physiome in any publication for which this software is used and send one reprint to the address given below:
The National Simulation Resource, Director J. B. Bassingthwaighte, Department of Bioengineering, University of Washington, Seattle WA 98195-5061.
Model development and archiving support at https://www.imagwiki.nibib.nih.gov/physiome provided by the following grants: NIH U01HL122199 Analyzing the Cardiac Power Grid, 09/15/2015 - 05/31/2020, NIH/NIBIB BE08407 Software Integration, JSim and SBW 6/1/09-5/31/13; NIH/NHLBI T15 HL88516-01 Modeling for Heart, Lung and Blood: From Cell to Organ, 4/1/07-3/31/11; NSF BES-0506477 Adaptive Multi-Scale Model Simulation, 8/15/05-7/31/08; NIH/NHLBI R01 HL073598 Core 3: 3D Imaging and Computer Modeling of the Respiratory Tract, 9/1/04-8/31/09; as well as prior support from NIH/NCRR P41 RR01243 Simulation Resource in Circulatory Mass Transport and Exchange, 12/1/1980-11/30/01 and NIH/NIBIB R01 EB001973 JSim: A Simulation Analysis Platform, 3/1/02-2/28/07.

