Introduction
In spite of the spectacular progress in molecular biology in recent years, the main challenge in biological and health sciences lies in the proper quantitative interpretation of carefully designed experiments. This asks for engineering strategies in the design of experiments and a rigorous theoretical basis of their interpretation. Endeavors of this kind are necessary to quantitatively describe and predict metabolic processes, hormonal activities and the pharmacological and toxic properties of drugs. They can contribute to a clearer picture of physiological functions and are also essential for the design of diagnostic procedures based on functonal imaging.
Actual rates in a given situation may be quite different from what may be expected from studying isolated systems. Therefore, in order to obtain a realistic idea of such rates, experiments have to be performed in intact cells, intact organs, or intact organisms.
For an intact organ, the simplest approach is one of mass balance where the net rates of the material fluxes entering and leaving the system under consideration are derived from concentration differences across the organ. This approach is futile if more than one pathway is involved in the metabolism of a given substrate, or if movement of substances between and within cells or chemical or enzymatic reactions are bi-directional. For example, lactate entering the liver may be either transformed into glucose or oxidised via the tricarboxylate cycle, while at the same time, lactate is produced by glycolysis. One might also wish to differentiate between the roles of individual sequential and/or parallel processes. The most powerful and convenient tool for this goal has been found to be the use of isotopically labeled substrates.
Application of radioactive tracers for the elucidation of biochemical pathways has a long tradition. However, the quantitative evaluation of the rate of individual steps in a pathway is a challenging task. The conventional approach consists in administrating a radioactively labelled substrate and evaluating the specific activity of products and possibly metabolic intermediates after a steady state has been achieved. Under favorable circumstances, this approach can yield flux rates for individual metabolic reactions; in many cases, however, a reliable evaluation of this kind of data is not possible. Moreover, measurements of steady-state tracer kinetics will not provide information on the sizes of metabolic pools.
Compartmental System Analysis and its Limitations
A more demanding approach involves the use of compartmental analysis, consisting in the application of tracer material as a bolus or as a step infusion, and the subsequent analysis of timed samples of the labelled substrate as well as of products and intermediates. It is further assumed that the tracer distributes in a small number of discrete compartments, and that within each of these, it distributes evenly, with the same concentration throughout the compartment. A convenient tool for modeling compartmental systems is the SAAM II software system.
In mathematical terms, compartmental analysis uses systems of ordinary differential equations. This is adequate in cases where the events under study are considerably slower than convection or diffusion, or where there is no flow, as is the case in many experimental systems involving isolated cell preparations. In many cases, however, membrane transport or metabolism is rapid enough to cause concentration gradients within the capillary bed, especially with substrates of intermediary metabolism where flux rates may become substantial. Therefore, previous models representing the organ as a single well-mixed compartment have to be replaced by distributed-in-space models. Rapid metabolism or membrane permeation interacts with blood flow in such a way that their contribution cannot be separated in a simple fashion. Under such circumstances, the classical form of compartmental analysis will fail; instead, a distributed-in-space concept has to be used, involving systems of partial differential equations.
The Multiple Indicator-Dilution Technique
The theoretical and experimental basis for the understanding of these phenomena has been laid by the development of the multiple indicator-dilution technique. Its principle is the injection of the substance to be examined into the artery of the organ (or the portal vein in the case of the liver), usually in radioactively labeled form. The label appearing in the venous perfusate is then analyzed as a function of time (outflow profiles). Simultaneously, non-interacting substances (“reference indicators”) are injected in order to gain information on the influence of blood flow on the outflow profiles. The rates of uptake, release, and metabolic transformation of the investigated substances are then obtained by comparison of their outflow profiles with those of the reference substances.
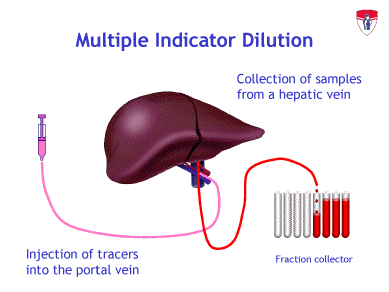
Originally, the multiple indicator-dilution technique had been used for studying the transport of metabolically inert materials, such as erythrocytes, water, potassium, or albumin. It has been successfully applied to the characterization of the transport of such substances by blood flow and transport across membranes in intact organs. For the liver, the most important work has been done by Goresky’s group.

Using the multiple indicator-dilution technique, metabolic events have mainly been considered in cases where metabolism is represented exclusively by irreversible removal of the substrate. More complex situations have been successfully analyzed, including the binding of monohydric alcohols to cytosolic alcohol dehydrogenase, the occurrence of an additional barrier due to the endothelium in heart capillaries and the involvement of erythrocytes in the carriage of the labelled substrate.
Examples
Back to Goresky Modeling of transport and metabolism tutorial
- Flow limited model
- Simple elimination with flow-limited distribution
- Barrier limited model
- Linear flow coupling
- Two-barrier model
- Red cell model
References
- Charnick SB, Kawai R, Nedelman JR, Lemaire M, Niederberger W and Sato H. Perspectives in pharmacokinetics. Physiologically based pharmacokinetic modeling as a tool for drug development. J Pharmacokinet Biopharm 23: 217-229, 1995
- Kelleher JK. Gluconeogenesis from labeled carbon: estimating isotope dilution. Am J Physiol 250: E296-E305, 1986.
- Müllhofer G, Schwab A, Müller C, Von Stetten C and Gruber E. Carbon-14 tracer studies in rat-liver perfusion experiments under conditions of gluconeogenesis from lactate and pyruvate. Eur J Biochem 75: 319-330, 1977.
- Jacquez JA. Compartmental Analysis in Biology and Medicine. Ann Arbor: The University of Michigan Press, 1986.
- (Unfinished) Catheter correction for Indicator dilution studies.
Author: Andreas J. Schwab (andreas.schwab@mcgill.ca)
Posted by: BEJ
Acknowledgements
Please cite https://www.imagwiki.nibib.nih.gov/physiome in any publication for which this software is used and send a copy to:
The National Simulation Resource, Director J. B. Bassingthwaighte, Department of Bioengineering, University of Washington, Seattle WA 98195-5061.
Model development and archiving support at https://www.imagwiki.nibib.nih.gov/physiome provided by the following grants: NIH U01HL122199 Analyzing the Cardiac Power Grid, 09/15/2015 - 05/31/2020, NIH/NIBIB BE08407 Software Integration, JSim and SBW 6/1/09-5/31/13; NIH/NHLBI T15 HL88516-01 Modeling for Heart, Lung and Blood: From Cell to Organ, 4/1/07-3/31/11; NSF BES-0506477 Adaptive Multi-Scale Model Simulation, 8/15/05-7/31/08; NIH/NHLBI R01 HL073598 Core 3: 3D Imaging and Computer Modeling of the Respiratory Tract, 9/1/04-8/31/09; as well as prior support from NIH/NCRR P41 RR01243 Simulation Resource in Circulatory Mass Transport and Exchange, 12/1/1980-11/30/01 and NIH/NIBIB R01 EB001973 JSim: A Simulation Analysis Platform, 3/1/02-2/28/07.

