Multiscale Modeling of Immune Dynamics
To join this working group please complete this form
Announcing the 2026 OpenVT Reproducible Agent-Based Multicellular Model Competition promoting FAIR, reproducible, and reusable multicellular simulations. Models must be published in a refereed journal on or after January 1, 2024 and publicly distributed.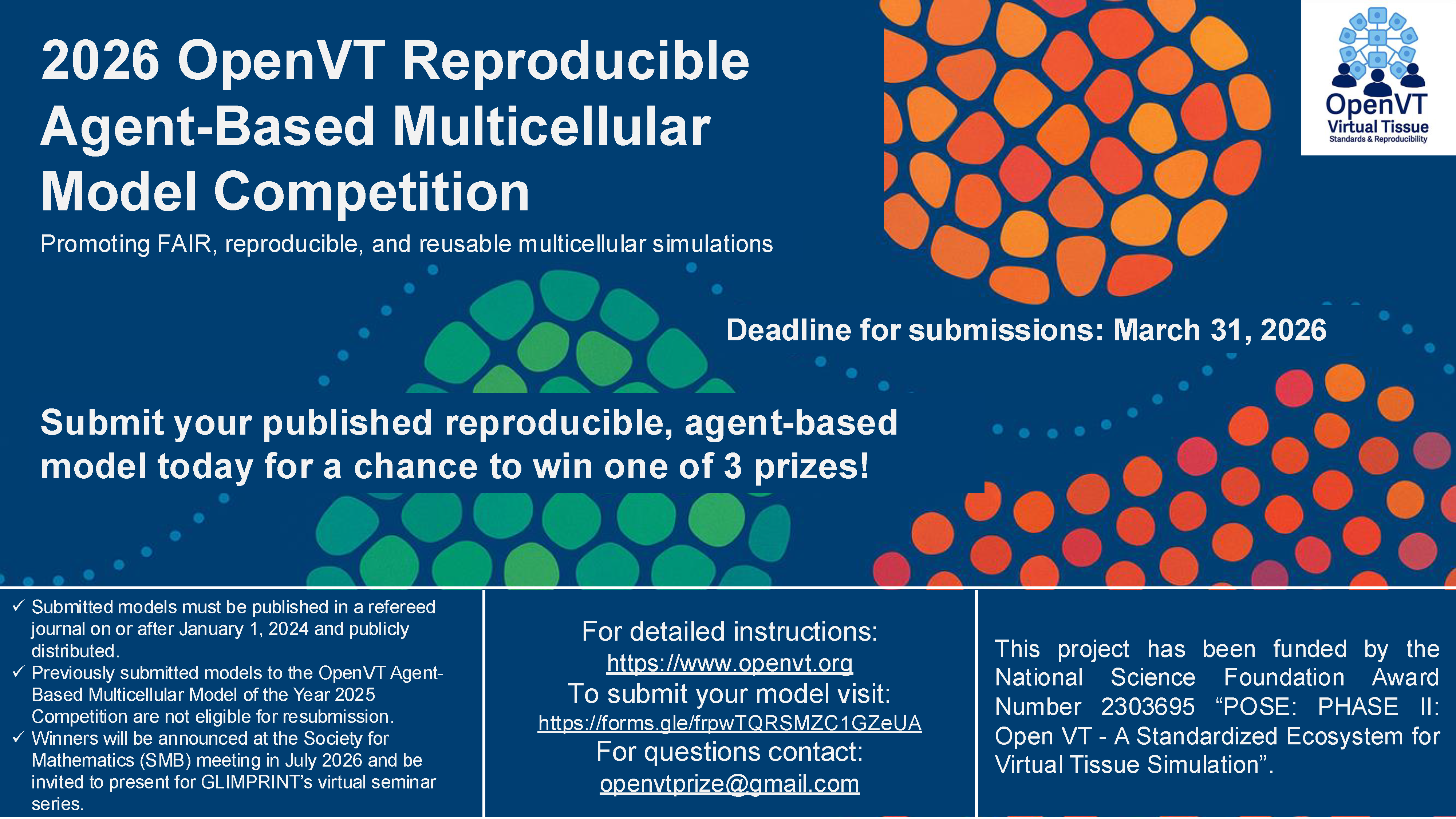
Extended Deadline for submission: March 31, 2026
For detailed instructions: https://www.openvt.org
To submit your model visit: https://forms.gle/frpwTQRSMZC1GZeUA
For questions contact: openvtprize@gmail.com
Winners will be invited to present in the GLIMPRINT’s virtual seminar series and at the Society for Mathematical Biology Annual Meeting (Joint with ECMTB) in Graz, Austria (13-17 July 2026). However, attending the meeting is not required to submit or to win.
Viral Pandemics WG Seminar Series
Our seminar series is now part of a consortium of modeling communities;
IMOBio, Integrative Multistage Open Biology Community.
February 26, 2026 at 10 AM US Eastern time
Andy Atkeson, UCLA
"An Economist's Perspective on Behavior and the Dynamics of Epidemics"
I use a model of private and public behavior to mitigate disease transmission during the COVID pandemic over the past year in the United States to address two questions: What dynamics of infections and deaths should we expect to see from a pandemic? and What are our options for mitigating the impact of a pandemic on public health? Economists posit that endogenous changes in behavior will drive the effective reproduction number of a disease towards one. For COVID, turns what would be a short and extremely sharp epidemic into a long, drawn out one. This assumption alters model implications for the purpose of public health interventions. For COVID, I calculate that, absent early eradication of the disease, non-pharmaceutical interventions alone suffer from rapidly diminishing returns in improving long-run health outcomes. In contrast, when these interventions are combined with a technological intervention such as vaccines, they can have a significant impact on public health outcomes. Many economists argued that, in both cases, more carefully targeted interventions could have attained similar health outcomes at significantly lower economic costs.
Please feel free to forward this invitation to anyone you think might be interested.
Please register at
https://iu.zoom.us/meeting/register/tZYqd-2srD8tGtCXDem4Cka08rBz5fDW0EQR#/registration
If you are interested in presenting a talk, please let us know.
Furthermore, if you have suggestions for potential speakers please fill out a Suggested Invitees Google Form. We will do our very best to schedule your potential seminar or to invite speakers according to your suggestions as soon as possible.
A complete list of past seminars, with links to YouTube recordings and sides, is available on our Seminars page.
Upcoming Seminars:
March 12, 2026 :
- John Hickey, Duke. “Living Maps: Using Spatial Omics to Simulate Cellular Behavior in Silico”
March 19, 2026 :
- Shabman Reed, NIH.
April 6, 2026 :
- James Doran, University of Bath. “Equation Learning for multiscale models of infectious diseases”
For details of the past and future teleconferences, including YouTube videos and slides,
please visit this page.
You can access the entire set of Viral Pandemic WG seminars at our YouTube Playlist
(click the "Subscribe" button if you would like to be notified when new videos are posted)
IMAG's COVID-19 modeling and data resources page is here.
Three VP Wiki pages:
(1) Publications, (2) Random but Relevant Things and (3) Viral Pandemics WG Runnable Models Page
Recent Publications from group members:
- Böttcher L, Fonseca LL, Laubenbacher RC. Control of medical digital twins with artificial neural networks. Philos Trans A Math Phys Eng Sci. 2025 Mar 13;383(2292):20240228. doi: 10.1098/rsta.2024.0228. Epub 2025 Mar 13. PMID: 40078154; PMCID: PMC11904622.
- Fonseca LL, Böttcher L, Mehrad B, Laubenbacher RC. Optimal control of agent-based models via surrogate modeling. PLoS Comput Biol. 2025 Jan 14;21(1):e1012138. doi: 10.1371/journal.pcbi.1012138. PMID: 39808665; PMCID: PMC11790234.
- Niarakis A, Laubenbacher R, An G, et al. Immune digital twins for complex human pathologies: applications, limitations, and challenges. NPJ Syst Biol Appl. 2024 Nov 30;10(1):141. doi: 10.1038/s41540-024-00450-5. PMID: 39616158; PMCID: PMC11608242. https://pmc.ncbi.nlm.nih.gov/articles/PMC11608242/
- OPINION: Medical ‘Digital Twins’ Will Lead the Way to Personalized Medicine. Reinhard C. Laubenbacher. Scientific American, May 14, 2024. https://www.scientificamerican.com/article/how-digital-twin-technology-harnesses-biology-and-computing-to-power/
- Gary An, Chase Cockrell. A design specification for Critical Illness Digital Twins to cure sepsis: responding to the National Academies of Sciences, Engineering and Medicine Report: Foundational Research Gaps and Future Directions for Digital Twins. 8 May 2024. arXiv. https://doi.org/10.48550/arXiv.2405.05301.
- Reinhard Laubenbacher, Frederick Adler, Gary An, Filippo Castiglione, Stephen Eubank, Luis L. Fonseca, James Glazier, Tomáš Helikar, Marti Jett-Tilton, Denise Kirschner, Paul Macklin, Borna Mehrad, Bethany Moore, Virginia Pasour, Ilya Shmulevich, Amber Smith, Isabel Voigt, Thomas Yankeelov, Tjalf Ziemssen. Toward Mechanistic Digital Twins: Some Use Cases. Front. Digit. Health, Sec. Health Informatics, Volume 6 - 2024 | doi: 10.3389/fdgth.2024.1349595 . (Provisionally accepted https://www.frontiersin.org/journals/digital-health/articles/10.3389/fdgth.2024.1349595/abstract)
- Laubenbacher R, Adler F, An G, Castiglione F, Eubank S, Fonseca LL, Glazier J, Helikar T, Jett-Tilton M, Kirschner D, Macklin P, Mehrad B, Moore B, Pasour V, Shmulevich I, Smith A, Voigt I, Yankeelov TE, Ziemssen T. Forum on immune digital twins: a meeting report. NPJ Syst Biol Appl. 2024 Feb 16;10(1):19. doi: 10.1038/s41540-024-00345-5. PMID: 38365857; PMCID: PMC10873299. https://www.nature.com/articles/s41540-024-00345-5
Media about group members or the group's activities:
- James Glazier's NOKIA interview podcast "Building the Star Trek Med-Bay" is available here!
- Son's health issues fuel UNL researcher's interest in building virtual immune system, Omaha World-Herald, Feb 27, 2022. Newspaper article about Tomáš Helikar's work on digital twins of the immune system, with additional commentary from James Glazier and Reinhard Laubenbacher.
- Medical Digital Twins: a New Frontier, ACM News Feb 24, 2022. ACM news article about Reinhard Laubenbacher's recent paper.
Recent Publications from group members (2022-2023 only):
- National Academies of Sciences, Engineering, and Medicine, Opportunities and Challenges for Digital Twins in Biomedical Research: Proceedings of a Workshop—in Brief. Casola L, editor. Washington (DC): National Academies Press (US); 2023 Jun 12. PMID: 37339242. https://pubmed.ncbi.nlm.nih.gov/37339242/
- Cockrell C, Laria D, An G. Preparing for the next pandemic: Simulation-based deep reinforcementlearning to discover and test multimodal control of systemic inflammation using repurposed immunomodulatory agents. Front. Immunol., 21 Nov. 2022 Sec. Systems Immunology https://doi.org/10.3389/fimmu.2022.995395
- Mochan, Ericka, T. J. Sego, and Bard Ermentrout. Age-Related Changes to the Immune System Exacerbate the Inflammatory Response to Pandemic H1N1 Infection. Bulletin of mathematical biology 84, no. 8 (2022): 1-24. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC9278316/
- Ferrari Gianlupi, Juliano, Tarunendu Mapder, T. J. Sego, James P. Sluka, Sara K. Quinney, Morgan Craig, Robert E. Stratford Jr, and James A. Glazier. Multiscale Model of Antiviral Timing, Potency, and Heterogeneity Effects on an Epithelial Tissue Patch Infected by SARS-CoV-2. Viruses 14, no. 3 (2022): 605. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8953050/
- Sego, T. J., Ericka D. Mochan, G. Bard Ermentrout, and James A. Glazier. A multiscale multicellular spatiotemporal model of local influenza infection and immune response. Journal of Theoretical Biology 532 (2022): 110918. https://www.ncbi.nlm.nih.gov/https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8478073/pmc/articles/PMC8478073/
- Böttcher L, Antulov-Fantulin N, Asikis T. AI Pontryagin or how artificial neural networks learn to control dynamical systems. Nat Commun. 2022 Jan 17;13(1):333. doi: 10.1038/s41467-021-27590-0. PMID: 35039488; PMCID: PMC8763915.
- Böttcher L, and Asikis T. Near-optimal control of dynamical systems with neural ordinary differential equations. Machine Learning: Science and Technology. Accepted 16 September 2022. Available online here.
- Waites W, Cavaliere M, Danos V, Datta R, Eggo RM, Hallett TB, Manheim D, Panovska-Griffiths J, Russell TW, Zarnitsyna VI. Compositional modelling of immune response and virus transmission dynamics. Philos Trans A Math Phys Eng Sci. 2022 Oct 3;380(2233):20210307. doi: 10.1098/rsta.2021.0307. Epub 2022 Aug 15. PMID: 35965463.
- Gary An and Chase Cockrell. Drug Development Digital Twins for Drug Discovery, Testing and Repurposing: A Schema for Requirements and Development. Front. Syst. Biol., 20 June 2022, Sec. Translational Systems Biology and In Silico Trials https://doi.org/10.3389/fsysb.2022.928387
- Darquenne C, Borojeni AAT, Colebank MJ, Forest MG, Madas BG, Tawhai M, Jiang Y. Aerosol Transport Modeling: The Key Link Between Lung Infections of Individuals and Populations. Front Physiol. 2022 Jun 20;13:923945. doi: 10.3389/fphys.2022.923945. PMID: 35795643; PMCID: PMC9251577. https://doi.org/10.3389/fphys.2022.923945
- Laubenbacher R, Niarakis A, Helikar T, An G, Shapiro B, Malik-Sheriff RS, Sego TJ, Knapp A, Macklin P, Glazier JA. Building digital twins of the human immune system: toward a roadmap NPJ Digit Med. 2022 May 20;5(1):64. doi: 10.1038/s41746-022-00610-z. PMID: 35595830; PMCID: PMC9122990.
- JM Sasso, BJB Ambrose, R Tenchov, RS Datta, MT Basel, RK DeLong, QA Zhou. The Progress and Promise of RNA Medicine─An Arsenal of Targeted Treatments. J Med Chem. 2022 May 9. doi: 10.1021/acs.jmedchem.2c00024. Epub ahead of print. PMID: 35533054.
- Dale Larie, Gary An, Robert Chase Cockrell. Preparing for the next COVID: Deep Reinforcement Learning trained Artificial Intelligence discovery of multi-modal immunomodulatory control of systemic inflammation in the absence of effective anti-microbials. http://dx.doi.org/10.1101/2022.02.17.480940
- An G. Specialty Grand Challenge: What it will take to cross the Valley of Death: Translational Systems Biology, “True” Precision Medicine, Medical Digital Twins, Artificial Intelligence and In Silico Clinical trials. https://www.frontiersin.org/articles/10.3389/fsysb.2022.901159/full
- Karr J, Malik-Sheriff RS, Osborne J, Gonzalez-Parra G, Forgoston E, Bowness R, Liu Y, Thompson R, Garira W, Barhak J, Rice J, Torres M, Dobrovolny HM, Tang T, Waites W, Glazier JA, Faeder JR and Kulesza A (2022) Model Integration in Computational Biology: The Role of Reproducibility, Credibility and Utility. Front. Syst. Biol. 2:822606. doi: 10.3389/fsysb.2022.822606 https://www.frontiersin.org/articles/10.3389/fsysb.2022.822606/full
- Larie D, An G, Cockrell RC. Preparing for the next COVID: Deep Reinforcement Learning trained Artificial Intelligence discovery of multi-modal immunomodulatory control of systemic inflammation in the absence of effective anti-microbials. Preprint, Feb. 18, 2022. https://www.biorxiv.org/content/10.1101/2022.02.17.480940v1.
Co-Leads:
James A. Glazier, PhD, Indiana University, JAGlazier@gmail.com
Tomas Helikar, PhD, University of Nebraska-Lincoln, thelikar2@unl.edu
Rationale: The ongoing COVID-19 pandemic has provided a striking example of the real-world importance of mathematical and computational modeling. Epidemiological simulations have become key technologies for optimizing responses by clinicians and policy makers around the world. Because epidemiological models had already been developed and validated for other infectious diseases before the appearance of SARS-CoV-2, these models were available for rapid repurposing when the COVID-19 crisis started. As a result, sophisticated epidemiological models of COVID-19 are informing healthcare professionals and public leaders as they decide on social restrictions or resource allocation.
However, once a patient is infected with SARS-CoV-2 (or another virus), current modeling technology is not sufficiently advanced to be of much help in assessing risk or guiding treatment. The spread of infection within the body and the immune response to respiratory and other viral pathogens is still poorly understood. The factors determining the beneficial (viral clearance) and harmful (cytokine storm) effects of immune response to COVID-19 are poorly understood. COVID-19 is also a very complex disease, with pathologies developing in organs beyond the sites of primary infection, and thus requiring understanding of the responses of multiple organ systems (especially the vasculature and blood) and their interactions. As the disease progresses over the course of weeks and months, coinfection between respiratory viruses is likely and its significance poorly understood. Therapies under consideration are also complex, with possible combination therapies combining phased dosages of novel and existing antivirals, pro-and anti-inflammatory drugs and antibodies. Such therapies will need to be personalized, and their combinatorial complexity makes evaluation with clinical studies which do not employ modeling for prioritization prohibitively time consuming and costly. The complex pattern of comorbidities to COVID-19 is suggestive but their significance is still unclear. Minority populations are at much greater risk both of being infected with SARS-CoV-2 and of dying from COVID-19, once infected. It is an urgent matter of health equity to understand and remediate these vulnerabilities. While the former (susceptibility) are primarily the subject of epidemiological models, the latter (death rates) are appropriate topics for the models the Working Group will consider. The COVID-19 epidemic has also brought modeling of infections into popular consciousness, education at all levels and policy making to an unprecedented degree. The development of models of human infection and response provide great opportunities for education and dissemination.
We currently lack platform tools for the evaluation of viral infections, their responses, and treatment opportunities like the epidemiology models that were available to customize for COVID-19. With partial exceptions for influenza and HIV/AIDS, few computational models attempt to collectively understand and harness the key drivers of infection progression for prognosis and optimal design of interventions. Even the most sophisticated current models do not usually include the details of immune response and inter-organ interactions that seem critical to COVID-19 and will be essential if the models are to serve as a platform for response to future viruses. Another obstacle to using modeling as a guide to treatment is the relative lack of ability to personalize such models using readily available data, such as lung CT scans, immune profiles from repeated blood draws, or comorbidities and other information from patient health records. This personalization of predefined and calibrated models is key, as COVID-19 has shown with its unpredictability of patient response. Added challenges are a better understanding of host-pathogen interactions, and the connection between the population and patient-level scales. These and other challenges make clear that the determination of an effective response to viral pandemics is a multiscale many-faceted problem whose solution has to rely on multiscale mathematical and computational models. The IMAG community is ideally positioned to lead an initiative to develop and help execute a strategy for developing and applying this technology.
Focus and structure of the working group: The community of modelers developing epidemiological and population-scale models is already extensive and well-integrated, in part due to the NIGMS MIDAS program. Within-host modeling of viral pathogens is much more limited. Therefore, the working group will initially focus on within-host scales, in particular the complex interactions between viral infection, host physiology, and the immune system. A main long-term deliverable of the working group will be an overall strategy for a coordinated multi-scale modeling effort which becomes a customizable translational technological platform for rapidly creating improved personalized prognoses and therapies in response to emerging viral pandemics. It will also include a plan on how to mobilize and coordinate the modeling community to support this effort.
The working group will be organized as follows:
A steering group, consisting of approximately 20 scientists, including modelers, data scientists, experimental and clinical domain experts, such as immunologists and virologists and representatives of affected communities and potential tool users, with special emphasis on the effects of the disease on disproportionately affected populations.
The steering group and subgroups will contain scientists from academia, the private sector, and government, also relying heavily on the members of the IMAG working groups. The working group will be widely advertised, and membership is open to all scientists. In addition, the co-leads will proactively invite scientists to join the steering group.
Member List is available here.
Subgroup pages
- Innate and Adaptive Immune Response
- Host-pathogen Interactions
- Tissue Damage and Recovery
- Viral Transport and Modes of Entry
- Viral Replication and Release
- Viral Evolution
- Vaccine Development
- Drug Development
- Modeling Individual Responses to Disease and Treatment
- Modeling Decontamination of Surfaces/Materials
- Machine Learning for Health Monitoring
- Heart and Vasculature
- Kidney and Liver
- Comorbidities
- Clinical and Experimental Data for Model Construction and Validation
- AI-based Data Processing, Heterogeneous Data Fusion
- Infection in Experimental Models
- Infection in zoonotic reservoir animals and Interspecies transmission
- Coinfection and/or Other Pathogens (Virus-Virus, Bacterial, Eukaryotic)
- Emerging and Reemerging Diseases
- Lung, Pulmonary Circulation and Aerosol Transport
- Integration
- Knowledge Acquisition and Modeling
- Building immune digital twins
- Model Reproducibility, Credibility and Standardization
- Dissemination and Outreach
- Communication to Funding Agencies
- TEMPLATE for subgroup pages
Initial list of deliverables and goals:
Six months:
Advertise for and recruit members as widely as possible.
Establishment of the steering group and subgroups with all needed areas of expertise, as well as an effective communication structure, including regular meetings and information sharing resources
A report that includes a map of the main processes to be modeled, available models, available data, and main laboratories around the world doing related research
List of most important models, data, techniques that are still missing
Plan of how to integrate existing as well as future models
Plan for approaches to validation and uncertainty quantification when model components are developed simultaneously and independently
Article about the initiative for a medical audience, published in NEJM, JAMA, or similar journal
Article about the initiative for a modeling audience, published in Nature Digital Medicine, or similar journal
One year:
Refinement of the six-month deliverables
Virtual one-day workshop on the report and next steps
Two years:
Comprehensive strategic plan for the development of a “digital twin” for therapeutics for an individual patient suffering from a viral infection
Plan for training of computational scientists who want to contribute to the initiative
Plan for use of models in training of STEM students and the general population
First steps toward implementation of this plan, including the identification of challenges and required resources
Five years:
Well-established organization of an international integrated research community in modeling of viral pandemics across scales
Slack Channel: msm-working-group.slack.com
YouTube Channel: You can also access the entire set of Viral Pandemic WG seminars at our YouTube Playlist:
https://www.youtube.com/playlist?list=PLiEtieOeWbMKh9VcQoinSwODcSZKMTGat
(click the "Subscribe" button if you would like to be notified when new videos are posted)
However, the most compete list of seminars (along with slides) is our seminars page at https://www.imagwiki.nibib.nih.gov/content/msm-viral-pandemics-meetings.
Twitter: https://twitter.com/MsmViral
If you would like access to the above please contact us at mailto:viralpandemMSM@gmail.com or via the Twitter handle https://twitter.com/MsmViral.
The initial teleconference was Thursday October 22, 2020 at 3PM US Eastern Time. The meeting had 72 attendees. The recording of the first meeting via Zoom is available at:
https://drive.google.com/file/d/1GOHG0ZA0khngnp-DH8Z31-LaBkag4D42/view?usp=sharing
Meetings and Teleconferences
We are holding zoom meetings for the entire working group every Thursday afternoon at 10AM Eastern US time. The complete schedule of past and future meetings is here.
Handy Links
We have a page of handy links for activities related to the working group here.
This page, our top level page for the Viral Pandemics WG has the tiny URL https://tinyurl.com/hkr97vfe .
Our initial charge to the working groups is outlined below:
Based on the proposal submitted to the IMAG/MSM Steering Committee, the objectives for the working group as a whole and each of the subgroups are as follows:
Objectives for subgroups for the first 9 months:
- Develop a plan to achieve the objectives listed below. (Target delivery December 31, 2020)
- Identify people working in this area and include a paragraph on their work. (Target delivery February 28, 2021)
- Recruit members
- Assemble a directory of researchers with bibliography
- Identify other subgroups that you should coordinate with
- Prepare a white paper, approx. 5pp in length, excluding references that does the following: (Target delivery May 31, 2021)
- Describe the focus of the subgroup, the major open problems to address, and the role modeling can play
- Describe what models and data are available, and the extent of our biological knowledge, available experimental systems, etc.
- Describe what is needed to address these problems that does not exist yet: models, data, experimental approaches, etc.
- Outline any action items that could get us to solutions to these problems.
These white papers can form the basis of a collective publication on the topic of multiscale modeling and viral pandemics.
- Catalyze research projects through presentations, exchange of ideas, search for strategic opportunities. (Target delivery August 30, 2021)
Note: The list below only includes individuals that are members of both the working group and members of IMAG/MSM. For a complete list of all members of the working group please see the Member List is here.

